Multiplex CRISPR-Cas Assay for Rapid, Isothermal and Visual Detection of White Spot Syndrome Virus (WSSV) and Enterocytozoon hepatopenaei (EHP) in Penaeid Shrimp
- PMID: 39628369
- PMCID: PMC11837468
- DOI: 10.1111/jfd.14059
Multiplex CRISPR-Cas Assay for Rapid, Isothermal and Visual Detection of White Spot Syndrome Virus (WSSV) and Enterocytozoon hepatopenaei (EHP) in Penaeid Shrimp
Abstract
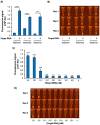
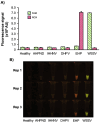
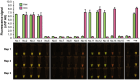
White spot syndrome virus (WSSV) and Enterocytozoon hepatopenaei (EHP) represent the most economically destructive pathogens in the current shrimp industry. WSSV causes white spot disease (WSD) responsible for rapid shrimp mortality, while EHP stunts growth and therefore reduces overall productivity. Despite the importance of timely disease detection, current diagnostic methods for WSSV and EHP are typically singleplex, and those offering multiplex detection face issues such as complexity, low field compatibility and/or low sensitivity. Here, we introduce an orthogonal, multiplex CRISPR-Cas assay for concomitant detection of WSSV and EHP. This method combines recombinase polymerase amplification (RPA) for target DNA enrichment with Cas12a and Cas13a enzymes for fluorescent detection. This assay produces distinct fluorescent colours for different diagnostic outcomes, allowing naked eye visualisation without ambiguity. Further validation reveals that the assay detects as few as 20 and 200 copies of target DNA from EHP and WSSV, respectively, while producing no false positives with DNA from other shrimp pathogens. Moreover, the assay excellently agrees with established PCR methods in evaluation of clinical samples. Requiring only 37°C and less than an hour to complete, multiplex CRISPR-Cas assay presents a promising tool for onsite diagnostics, offering high accuracy while saving time and resources.
Keywords: CRISPR diagnostics; Cas12a; Cas13a; Enterocytozoon hepatopenaei (EHP); penaeid shrimp; white spot syndrome virus (WSSV).
© 2024 The Author(s). Journal of Fish Diseases published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Multiple infections caused by white spot syndrome virus and Enterocytozoon hepatopenaei in pond-reared Penaeus vannamei in India and multiplex PCR for their simultaneous detection.J Fish Dis. 2019 Mar;42(3):447-454. doi: 10.1111/jfd.12956. Epub 2019 Jan 18. J Fish Dis. 2019. PMID: 30659620
-
CRISPR-Cas fluorescent cleavage assay coupled with recombinase polymerase amplification for sensitive and specific detection of Enterocytozoon hepatopenaei.Biotechnol Rep (Amst). 2020 Jun 6;27:e00485. doi: 10.1016/j.btre.2020.e00485. eCollection 2020 Sep. Biotechnol Rep (Amst). 2020. PMID: 32577410 Free PMC article.
-
Development of a PCR assay for the effective detection of Enterocytozoon hepatopenaei (EHP) and investigation of EHP prevalence in Shandong Province, China.J Invertebr Pathol. 2021 Sep;184:107653. doi: 10.1016/j.jip.2021.107653. Epub 2021 Aug 8. J Invertebr Pathol. 2021. PMID: 34371089
-
CRISPR-Cas assisted diagnostics of plant viruses and challenges.Virology. 2024 Sep;597:110160. doi: 10.1016/j.virol.2024.110160. Epub 2024 Jun 26. Virology. 2024. PMID: 38955083 Review.
-
Host-Parasite Interactions and Integrated Management Strategies for Ecytonucleospora Hepatopenaei Infection in Shrimp.Acta Parasitol. 2025 Mar 6;70(2):67. doi: 10.1007/s11686-025-01007-0. Acta Parasitol. 2025. PMID: 40050501 Review.
Cited by
-
Shrimp White Spot Viral Infections Are Attenuated by Organic Acids by Regulating the Expression of HO-1 Oxygenase and β-1,3-Glucan-Binding Protein.Antioxidants (Basel). 2025 Jan 14;14(1):89. doi: 10.3390/antiox14010089. Antioxidants (Basel). 2025. PMID: 39857423 Free PMC article.
References
-
- Babu, B. , Sathiyaraj G., Mandal A., et al. 2021. “Surveillance of Disease Incidence in Shrimp Farms Located in the East Coastal Region of India and In Vitro Antibacterial Efficacy of Probiotics Against Vibrio parahaemolyticus .” Journal of Invertebrate Pathology 179: 107536. 10.1016/J.JIP.2021.107536. - DOI - PubMed
-
- Bandeira, J. d. T. , de Morais R. S. M. M., Silva R. P. P. E., Mendes E. S., da Silva S. M. B. C., and Santos F. L. d.. 2019. “First Report of White Spot Syndrome Virus in Wild Crustaceans and Mollusks in the Paraíba River, Brazil.” Aquaculture Research 50, no. 2: 680–684. 10.1111/are.13949. - DOI
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

