The impact of a polyphenol-rich supplement on epigenetic and cellular markers of immune age: a pilot clinical study
- PMID: 39628466
- PMCID: PMC11612904
- DOI: 10.3389/fnut.2024.1474597
The impact of a polyphenol-rich supplement on epigenetic and cellular markers of immune age: a pilot clinical study
Abstract
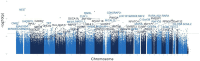
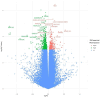
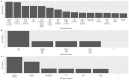
Age-related alterations in immune function are believed to increase risk for a host of age-related diseases leading to premature death and disability. Programming of the immune system by diet, lifestyle, and environmental factors occurs across the lifespan and influences both makeup and function of the immune system, including immunometabolism. This programming is believed to act in large part through epigenetic modification. Among dietary components that affect this process, polyphenols may play an outsized role. Polyphenols are a widely distributed group of plant nutrients consumed by humans. Certain foods possess distinctive and relatively higher levels of these compounds. One such food is Tartary buckwheat (fagopyrum tataricum), an ancient seed historically prized for its health benefits. It is suggested that the specific composition of polyphenols found in foods like Tartary buckwheat may lead to a unique impact on immunometabolic physiological pathways that could be interrogated through epigenetic analyses. The objective of this study was to investigate the epigenetic effects on peripheral immune cells in healthy individuals of a standardized polyphenol concentrate based on naturally occurring nutrients in Tartary buckwheat. This pilot clinical trial tested the effects of consuming 90 days of this concentrate in 50 healthy male (40%) and female (60%) participants aged 18-85 years using epigenetic age clocks and deconvolution methods. Analysis revealed significant intervention-related changes in multiple epigenetic age clocks and immune markers as well as population-wide alterations in gene ontology (GO) pathways related to longevity and immunity. This study provides previously unidentified insights into the immune, longevity and epigenetic effects of consumption of polyphenol-rich plants and generates additional support for health interventions built around historically consumed plants like Tartary buckwheat while offering compelling opportunities for additional research.
Clinical trial registration: ClinicalTrials.gov, Identifier: NCT05234203.
Keywords: Tartary buckwheat; aging; diet and nutrition; epigenetic clocks; epigenome-wide association study; food-is-medicine; immunity; polyphenols.
Copyright © 2024 Perlmutter, Bland, Chandra, Malani, Smith, Mendez and Dwaraka.
Conflict of interest statement
JSB and AP are employees of Big Bold Health. RS, VBD, and TLM are employees of TruDiagnostic. SM and AC are consultants to Big Bold Health.
Figures



References
Associated data
LinkOut - more resources
Full Text Sources
Medical

