Distinct effects of sacituzumab govitecan and berzosertib on DNA damage response in ovarian cancer
- PMID: 39628575
- PMCID: PMC11613210
- DOI: 10.1016/j.isci.2024.111283
Distinct effects of sacituzumab govitecan and berzosertib on DNA damage response in ovarian cancer
Abstract
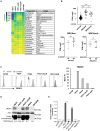
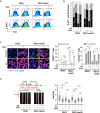
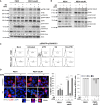
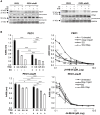
Antibody-drug conjugates (ADCs) have become an important class of anticancer drugs in solid tumors including drug-resistant gynecologic malignancies. TROP2 is a cell surface antigen that is highly expressed in ovarian carcinoma (OC) but minimally expressed in normal ovarian tissues. In this study, we aimed to identify how TROP2-specific ADC, sacituzumab govitecan (SG), modulates DNA damage response pathways in drug-resistant OC. We found that SG induces G2/M arrest, increases RPA1 foci, and decreases replication fork speed, resulting in replication stress in TROP2-positive cells while these were less evident in TROP2-negative cells. In OC in vitro and in vivo models, SN-38 sensitivity and TROP2 expression play key roles in response to either ATR inhibitor or SG alone, or in combination. Additionally, inhibition of translesion DNA synthesis enhances SG and PARP inhibitor (PARPi) sensitivity in PARPi-resistant OC cells. These findings provide mechanistic insights for clinical development of SG in drug-resistant OC.
Keywords: Cancer; Molecular biology; Oncology.
Conflict of interest statement
T.M.C. is a paid contract employee of Gilead Sciences, Inc. J.M.L. has research grant funding from AstraZeneca and Acrivon Therapeutics (paid to institution) and is on the Scientific Advisory Board of Acrivon Therapeutics and Genentech (unpaid). All other authors declare no conflict of interest.
Figures




References
-
- Konstantinopoulos P.A., Spentzos D., Karlan B.Y., Taniguchi T., Fountzilas E., Francoeur N., Levine D.A., Cannistra S.A. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol. 2010;28:3555–3561. doi: 10.1200/JCO.2009.27.5719. - DOI - PMC - PubMed
-
- Brill E., Yokoyama T., Nair J., Yu M., Ahn Y.R., Lee J.M. Prexasertib, a cell cycle checkpoint kinases 1 and 2 inhibitor, increases in vitro toxicity of PARP inhibition by preventing Rad51 foci formation in BRCA wild type high-grade serous ovarian cancer. Oncotarget. 2017;8:111026–111040. doi: 10.18632/oncotarget.22195. - DOI - PMC - PubMed
-
- Cheng X., Li J., Tanaka K., Majumder U., Milinichik A.Z., Verdi A.C., Maddage C.J., Rybinski K.A., Fernando S., Fernando D., et al. MORAb-202, an antibody--drug conjugate utilizing humanized anti-human FR$∖alpha$ farletuzumab and the microtubule-targeting agent eribulin, has potent antitumor activity. Mol. Cancer Therapeut. 2018;17:2665–2675. doi: 10.1158/1535-7163.MCT-17-1215. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

