Oxidized mRNA Lipid Nanoparticles for In Situ Chimeric Antigen Receptor Monocyte Engineering
- PMID: 39628840
- PMCID: PMC11611297
- DOI: 10.1002/adfm.202312038
Oxidized mRNA Lipid Nanoparticles for In Situ Chimeric Antigen Receptor Monocyte Engineering
Abstract
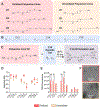
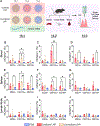
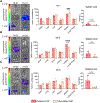
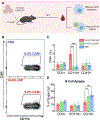
Chimeric antigen receptor (CAR) monocyte and macrophage therapies are promising solid tumor immunotherapies that can overcome the challenges facing conventional CAR T cell therapy. mRNA lipid nanoparticles (mRNA-LNPs) offer a viable platform for in situ engineering of CAR monocytes with transient and tunable CAR expression to reduce off-tumor toxicity and streamline cell manufacturing. However, identifying LNPs with monocyte tropism and intracellular delivery potency is difficult using traditional screening techniques. Here, ionizable lipid design and high-throughput in vivo screening are utilized to identify a new class of oxidized LNPs with innate tropism and mRNA delivery to monocytes. A library of oxidized (oLNPs) and unoxidized LNPs (uLNPs) is synthesized to evaluate mRNA delivery to immune cells. oLNPs demonstrate notable differences in morphology, ionization energy, and pKa, therefore enhancing delivery to human macrophages, but not T cells. Subsequently, in vivo library screening with DNA barcodes identifies an oLNP formulation, C14-O2, with innate tropism to monocytes. In a proof-of-concept study, the C14-O2 LNP is used to engineer functional CD19-CAR monocytes in situ for robust B cell aplasia (45%) in healthy mice. This work highlights the utility of oxidized LNPs as a promising platform for engineering CAR macrophages/monocytes for solid tumor CAR monocyte therapy.
Figures



References
-
- CAR T Cells: Engineering Immune Cells to Treat Cancer - NCI. https://www.cancer.gov/about-cancer/treatment/research/car-t-cells (2013).
Grants and funding
LinkOut - more resources
Full Text Sources
