AnACor2.0: a GPU-accelerated open-source software package for analytical absorption corrections in X-ray crystallography
- PMID: 39628882
- PMCID: PMC11611279
- DOI: 10.1107/S1600576724009506
AnACor2.0: a GPU-accelerated open-source software package for analytical absorption corrections in X-ray crystallography
Abstract
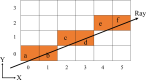
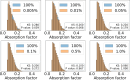
Analytical absorption corrections are employed in scaling diffraction data for highly absorbing samples, such as those used in long-wavelength crystallography, where empirical corrections pose a challenge. AnACor2.0 is an accelerated software package developed to calculate analytical absorption corrections. It accomplishes this by ray-tracing the paths of diffracted X-rays through a voxelized 3D model of the sample. Due to the computationally intensive nature of ray-tracing, the calculation of analytical absorption corrections for a given sample can be time consuming. Three experimental datasets (insulin at λ = 3.10 Å, thermolysin at λ = 3.53 Å and thaumatin at λ = 4.13 Å) were processed to investigate the effectiveness of the accelerated methods in AnACor2.0. These methods demonstrated a maximum reduction in execution time of up to 175× compared with previous methods. As a result, the absorption factor calculation for the insulin dataset can now be completed in less than 10 s. These acceleration methods combine sampling, which evaluates subsets of crystal voxels, with modifications to standard ray-tracing. The bisection method is used to find path lengths, reducing the complexity from O(n) to O(log2 n). The gridding method involves calculating a regular grid of diffraction paths and using interpolation to find an absorption correction for a specific reflection. Additionally, optimized and specifically designed CUDA implementations for NVIDIA GPUs are utilized to enhance performance. Evaluation of these methods using simulated and real datasets demonstrates that systematic sampling of the 3D model provides consistently accurate results with minimal variance across different sampling ratios. The mean difference of absorption factors from the full calculation (without sampling) is at most 2%. Additionally, the anomalous peak heights of sulfur atoms in the Fourier map show a mean difference of only 1% compared with the full calculation. This research refines and accelerates the process of analytical absorption corrections, introducing innovative sampling and computational techniques that significantly enhance efficiency while maintaining accurate results.
Keywords: CUDA acceleration; absorption correction; long-wavelength crystallography; ray-tracing; software.
© Yishun Lu et al. 2024.
Figures







References
-
- Albrecht, G. (1939). Rev. Sci. Instrum.10, 221–222.
-
- Amanatides, J. & Woo, A. (1987). EG1987 Proceedings (Technical Papers). Eindhoven: Eurographics Association.
-
- Angel, R. J. (2004). J. Appl. Cryst.37, 486–492.
-
- Bruker (2016). SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
LinkOut - more resources
Full Text Sources
Research Materials
