Advances in the dynamic control of metabolic pathways in Saccharomyces cerevisiae
- PMID: 39628908
- PMCID: PMC11610979
- DOI: 10.1016/j.engmic.2023.100103
Advances in the dynamic control of metabolic pathways in Saccharomyces cerevisiae
Abstract
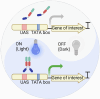
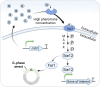
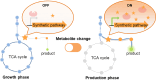
The metabolic engineering of Saccharomyces cerevisiae has great potential for enhancing the production of high-value chemicals and recombinant proteins. Recent studies have demonstrated the effectiveness of dynamic regulation as a strategy for optimizing metabolic flux and improving production efficiency. In this review, we provide an overview of recent advancements in the dynamic regulation of S. cerevisiae metabolism. Here, we focused on the successful utilization of transcription factor (TF)-based biosensors within the dynamic regulatory network of S. cerevisiae. These biosensors are responsive to a wide range of endogenous and exogenous signals, including chemical inducers, light, temperature, cell density, intracellular metabolites, and stress. Additionally, we explored the potential of omics tools for the discovery of novel responsive promoters and their roles in fine-tuning metabolic networks. We also provide an outlook on the development trends in this field.
Keywords: Dynamic control; Metabolic engineering; Response element; Transcription regulation.
© 2023 The Authors. Published by Elsevier B.V. on behalf of Shandong University.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Transcriptional regulatory proteins in central carbon metabolism of Pichia pastoris and Saccharomyces cerevisiae.Appl Microbiol Biotechnol. 2020 Sep;104(17):7273-7311. doi: 10.1007/s00253-020-10680-2. Epub 2020 Jul 10. Appl Microbiol Biotechnol. 2020. PMID: 32651601 Review.
-
Promoter Architecture and Promoter Engineering in Saccharomyces cerevisiae.Metabolites. 2020 Aug 6;10(8):320. doi: 10.3390/metabo10080320. Metabolites. 2020. PMID: 32781665 Free PMC article. Review.
-
Metabolic engineering of Saccharomyces cerevisiae for the synthesis of valuable chemicals.Crit Rev Biotechnol. 2024 Mar;44(2):163-190. doi: 10.1080/07388551.2022.2153008. Epub 2023 Jan 3. Crit Rev Biotechnol. 2024. PMID: 36596577 Review.
-
Dynamic pathway regulation: recent advances and methods of construction.Curr Opin Chem Biol. 2017 Dec;41:28-35. doi: 10.1016/j.cbpa.2017.10.004. Epub 2017 Oct 20. Curr Opin Chem Biol. 2017. PMID: 29059607 Review.
-
Metabolic engineering of Saccharomyces cerevisiae for glycerol utilization.FEMS Yeast Res. 2023 Jan 4;23:foad014. doi: 10.1093/femsyr/foad014. FEMS Yeast Res. 2023. PMID: 36869777 Review.
Cited by
-
The advances in creating Crabtree-negative Saccharomyces cerevisiae and the application for chemicals biosynthesis.FEMS Yeast Res. 2025 Jan 30;25:foaf014. doi: 10.1093/femsyr/foaf014. FEMS Yeast Res. 2025. PMID: 40121184 Free PMC article. Review.
-
Advances in synthesizing plant-derived isoflavones and their precursors with multiple pharmacological activities using engineered yeasts.Microb Cell Fact. 2025 Mar 29;24(1):75. doi: 10.1186/s12934-025-02692-2. Microb Cell Fact. 2025. PMID: 40155940 Free PMC article. Review.
-
Bulk and selective autophagy cooperate to remodel a fungal proteome in response to changing nutrient availability.bioRxiv [Preprint]. 2024 Sep 26:2024.09.24.614842. doi: 10.1101/2024.09.24.614842. bioRxiv. 2024. PMID: 39386609 Free PMC article. Preprint.
References
-
- Nandy S.K., Srivastava R.K. A review on sustainable yeast biotechnological processes and applications. Microbiol. Res. 2018;207:83–90. - PubMed
-
- Borodina I., Nielsen J. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 2014;9(5):609–620. - PubMed
-
- Kwak S., et al. Redirection of the glycolytic flux enhances isoprenoid production in Saccharomyces cerevisiae. Biotechnol. J. 2020;15(2) - PubMed
-
- Zhao Y., et al. Production of β-carotene in Saccharomyces cerevisiae through altering yeast lipid metabolism. Biotechnol. Bioeng. 2021;118(5):2043–2052. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

