Impact of camel milk lactoferrin peptides against breast cancer cells: in silico and in vitro study
- PMID: 39629082
- PMCID: PMC11612555
- DOI: 10.3389/fphar.2024.1425504
Impact of camel milk lactoferrin peptides against breast cancer cells: in silico and in vitro study
Abstract
Background and aims: Breast cancer remains a significant global health concern, necessitating the exploration of novel therapeutic strategies. Despite advancements in cancer therapeutics, effective treatments with minimal side effects remain elusive. Natural sources, such as camel milk, harbor bioactive compounds such as lactoferrin peptides, which hold promise as anticancer agents. This study investigated the potential of camel milk-derived lactoferrin peptides against breast cancer cells through a combined in silico and in vitro approach. By integrating computational modeling with experimental assays, we aimed to elucidate the anticancer mechanisms of these peptides and provide insights for their optimization as anticancer therapeutics.
Methods: In silico analysis involving pepetid design, and validation, then molecular docking and molecular dynamics (MD) simulations was used to explore peptide-protein interactions and stability. Peptides were synthesized and tested for anticancer activity using MTT assays on MCF-7 cells, with HDFa normal cells used as controls.
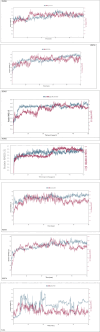
Results: Results of this study showed that camel milk-derived lactoferrin peptides, particularly PEP66, exhibited strong anticancer activity against MCF-7 breast cancer cells, with the lowest IC50 value (52.82 μg/mL) compared to other peptides. In silico molecular docking and dynamics simulations revealed that PEP66 formed stable interactions with key residues in the HER2 catalytic site, indicating its potential as an effective anticancer agent. The selectivity index (SI) of PEP66 (3.19) also suggested lower toxicity to normal cells compared to cancer cells, reinforcing its therapeutic potential. Hydrogen bonding analysis highlighted key residues involved in stabilizing peptide-protein complexes, while molecular dynamics simulations demonstrated the stability of these interactions over time. Notably, PEP66 exhibited the highest stability and formed significant interactions with essential residues in the HER2 catalytic site, suggesting its potential as an effective anticancer agent.
Conclusion: Camel milk-derived lactoferrin peptides show promise as anticancer agents against breast cancer cells. The multidisciplinary approach employed in this study provides valuable insights into the mechanisms underlying their activity, paving the way for rational design strategies to enhance their efficacy. Further experimental validation is warranted to validate the anticancer potential of these peptides and advance their development as novel therapeutic agents for breast cancer treatment.
Keywords: HDFa normal cells; HER2 protein; MCF-7 breast cancer cells; camel milk; in silico.
Copyright © 2024 Baothman, Ali, Alguridi, Hosawi, Konozy, Abu Zeid, Ahmad and Altayb.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Ali E. M., Elashkar A. A., El-Kassas H. Y., Salim E. I. (2018). Methotrexate loaded on magnetite iron nanoparticles coated with chitosan: biosynthesis, characterization, and impact on human breast cancer MCF-7 cell line. Int. J. Biol. Macromol. 120, 1170–1180. 10.1016/j.ijbiomac.2018.08.118 - DOI - PubMed
-
- Alkhulaifi M. M., Alosaimi M. M., Khan M. S., Tabrez S., Shaik G. M., Alokail M. S., et al. (2024). Assessment of broad-spectrum antimicrobial, antibiofilm, and anticancer potential of lactoferrin extracted from camel milk. Appl. Biochem. Biotechnol. 196, 1464–1480. 10.1007/s12010-023-04579-7 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

