Metagenomics insights into bacterial diversity and antibiotic resistome of the sewage in the city of Belém, Pará, Brazil
- PMID: 39629213
- PMCID: PMC11611572
- DOI: 10.3389/fmicb.2024.1466353
Metagenomics insights into bacterial diversity and antibiotic resistome of the sewage in the city of Belém, Pará, Brazil
Abstract
Introduction: The advancement of antimicrobial resistance is a significant public health issue today. With the spread of resistant bacterial strains in water resources, especially in urban sewage, metagenomic studies enable the investigation of the microbial composition and resistance genes present in these locations. This study characterized the bacterial community and antibiotic resistance genes in a sewage system that receives effluents from various sources through metagenomics.
Methods: One liter of surface water was collected at four points of a sewage channel, and after filtration, the total DNA was extracted and then sequenced on an NGS platform (Illumina® NextSeq). The sequenced data were trimmed, and the microbiome was predicted using the Kraken software, while the resistome was analyzed on the CARD webserver. All ecological and statistical analyses were performed using the. RStudio tool.
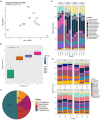
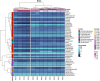
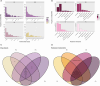
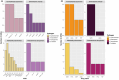
Results and discussion: The complete metagenome results showed a community with high diversity at the beginning and more restricted diversity at the end of the sampling, with a predominance of the phyla Bacteroidetes, Actinobacteria, Firmicutes, and Proteobacteria. Most species were considered pathogenic, with an emphasis on those belonging to the Enterobacteriaceae family. It was possible to identify bacterial groups of different threat levels to human health according to a report by the U.S. Centers for Disease Control and Prevention. The resistome analysis predominantly revealed genes that confer resistance to multiple drugs, followed by aminoglycosides and macrolides, with efflux pumps and drug inactivation being the most prevalent resistance mechanisms. This work was pioneering in characterizing resistance in a sanitary environment in the Amazon region and reinforces that sanitation measures for urban sewage are necessary to prevent the advancement of antibiotic resistance and the contamination of water resources, as evidenced by the process of eutrophication.
Keywords: antibiotic resistant bacteria; metagenome; microbial diversity; resistome; sewerage.
Copyright © 2024 Ramos, Júnior, Alegria, Vieira, Patroca, Cecília, Moreira and Nunes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China.Microbiome. 2017 Jul 19;5(1):84. doi: 10.1186/s40168-017-0298-y. Microbiome. 2017. PMID: 28724443 Free PMC article.
-
Population-based variations of a core resistome revealed by urban sewage metagenome surveillance.Environ Int. 2022 May;163:107185. doi: 10.1016/j.envint.2022.107185. Epub 2022 Mar 17. Environ Int. 2022. PMID: 35306253
-
The relationship between water quality and the microbial virulome and resistome in urban streams in Brazil.Environ Pollut. 2024 May 1;348:123849. doi: 10.1016/j.envpol.2024.123849. Epub 2024 Mar 22. Environ Pollut. 2024. PMID: 38522607
-
Metagenomics insights into the profiles of antibiotic resistome in combined sewage overflows from reads to metagenome assembly genomes.J Hazard Mater. 2022 May 5;429:128277. doi: 10.1016/j.jhazmat.2022.128277. Epub 2022 Jan 19. J Hazard Mater. 2022. PMID: 35074753
-
Profiles of Microbial Community and Antibiotic Resistome in Wild Tick Species.mSystems. 2022 Aug 30;7(4):e0003722. doi: 10.1128/msystems.00037-22. Epub 2022 Aug 1. mSystems. 2022. PMID: 35913190 Free PMC article. Review.
References
-
- Alam M. U., Ferdous S., Ercumen A., Lin A., Kamal A., Luies S. K., et al. . (2021). Effective treatment strategies for the removal of antibiotic-resistant Bacteria, antibiotic-resistance genes, and antibiotic residues in the effluent from wastewater treatment plants receiving municipal, hospital, and domestic wastewater: protocol for a systematic review. JMIR Res. Protoc. 10:e33365. doi: 10.2196/33365, PMID: - DOI - PMC - PubMed
-
- Al Salah D. M. M., Laffite A., Sivalingam P., Poté J. (2022). Occurrence of toxic metals and their selective pressure for antibiotic-resistant clinically relevant bacteria and antibiotic-resistant genes in river receiving systems under tropical conditions. Environ. Sci. Pollut. Res. Int. 29, 20530–20541. doi: 10.1007/s11356-021-17115-z, PMID: - DOI - PMC - PubMed
-
- Anderson M. J. (2017). Permutational Multivariate Analysis of Variance (PERMANOVA). In: Wiley StatsRef: Statistics Reference Online. Eds. Balakrishnan N., Colton T., Everitt B., Piegorsch W., Ruggeri F. and Teugels, J. L.
LinkOut - more resources
Full Text Sources

