Gonadal sex reversal at single-cell resolution in Znrf3-deficient mice
- PMID: 39629665
- PMCID: PMC11658683
- DOI: 10.1242/dev.202707
Gonadal sex reversal at single-cell resolution in Znrf3-deficient mice
Abstract
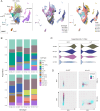
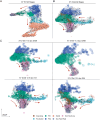
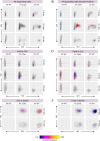
The role of anti-WNT ZNRF3 is central to determining gonadal fate: XY mice lacking functional ZNRF3 exhibit a highly variable gonadal sex reversal phenotype in the fetal period, characterised by appearance of ovarian tissue. To investigate this sex reversal further, we used single-cell RNA-seq to examine the transcriptomes of XY Znrf3-deficient gonads during the mouse sex-determining period. Analyses of cell trajectories in mutant gonads reveal the failure of pre-supporting cells to commit to the Sertoli cell fate, XY granulosa cell development, unstable commitment in those cells that reach the Sertoli path and enhanced contribution to a supporting-like cell fate. By developing a machine learning-based score for transcriptomic similarity to Sertoli and granulosa, we show pervasive disruption to acquisition of testicular cell fate in the mutant supporting cell lineage, with large numbers of cells co-expressing pro-Sertoli and pro-granulosa markers. These data reveal that loss of Znrf3 results in transcriptomic and cellular heterogeneity, with shifts in cellular sex identity that undermine a simple binary model in which mutant supporting cell precursors achieve either Sertoli or granulosa cell differentiation.
Keywords: Genetics of sex; Mouse; Ovotestis; Sex determination; Sex reversal; Testis; Znrf3.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


References
-
- Alles, J., Karaiskos, N., Praktiknjo, S. D., Grosswendt, S., Wahle, P., Ruffault, P. L., Ayoub, S., Schreyer, L., Boltengagen, A., Birchmeier, C., Zinzen, R., Kocks, C. and Rajewsky, N. (2017). Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 15, 44. 10.1186/s12915-017-0383-5 - DOI - PMC - PubMed
-
- Bogani, D., Siggers, P., Brixey, R., Warr, N., Beddow, S., Edwards, J., Williams, D., Wilhelm, D., Koopman, P., Flavell, R. A.et al. (2009). Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 7, e1000196. 10.1371/journal.pbio.1000196 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

