A genome-wide association study of European advanced cancer patients treated with opioids identifies regulatory variants on chromosome 20 associated with pain intensity
- PMID: 39629963
- PMCID: PMC11616469
- DOI: 10.1002/ejp.4764
A genome-wide association study of European advanced cancer patients treated with opioids identifies regulatory variants on chromosome 20 associated with pain intensity
Abstract
Background: Opioids in step III of the WHO analgesic ladder are the standard of care for treating cancer pain. However, a significant minority of patients do not benefit from therapy. Genetics might play a role in predisposing patients to a good or poor response to opioids. Here, we investigated this issue by conducting a genome-wide association study (GWAS).
Methods: We genotyped 2057 European advanced cancer patients treated with morphine, buprenorphine, fentanyl and oxycodone. We carried out a whole-genome regression model (using REGENIE software) between genotypes and the opioid response phenotype, defined as a numerical score measuring patient pain intensity.
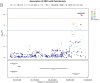
Results: The GWAS identified five non-coding variants on chromosome 20 with a p-value <5.0 × 10-8. For all of them, the minor allele was associated with lower pain intensity. These variants were intronic to the PCMTD2 gene and were 200 kbp downstream of OPRL1, the opioid related nociceptin receptor 1. Notably according to the eQTLGen database, these variants act as expression quantitative trait loci, modulating the expression mainly of PCMTD2 but also of OPRL1. Variants in the same chromosomal region were recently reported to be significantly associated with pain intensity in a GWAS conducted in subjects with different chronic pain conditions.
Conclusions: Our results support the role of genetics in the opioid response in advanced cancer patients. Further functional analyses are needed to understand the biological mechanism underlying the observed association and lead to the development of individualized pain treatment plans, ultimately improving the quality of life for cancer patients.
Significance statement: This genome-wide association study on European advanced cancer patients treated with opioids identifies novel regulatory variants on chromosome 20 (near PCMTD2 and OPRL1 genes) associated with pain intensity. These findings enhance our understanding of the genetic basis of opioid response, suggesting new potential markers for opioid efficacy. The study is a significant advancement in pharmacogenomics, providing a robust dataset and new insights into the genetic factors influencing pain intensity, which could lead to personalized cancer pain management.
© 2024 The Author(s). European Journal of Pain published by John Wiley & Sons Ltd on behalf of European Pain Federation ‐ EFIC ®.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Abascal, F. , Acosta, R. , Addleman, N. J. , Adrian, J. , Afzal, V. , Ai, R. , Aken, B. , Akiyama, J. A. , al Jammal, O. , Amrhein, H. , Anderson, S. M. , Andrews, G. R. , Antoshechkin, I. , Ardlie, K. G. , Armstrong, J. , Astley, M. , Banerjee, B. , Barkal, A. A. , Barnes, I. H. A. , … Ecker, J. R. (2020). Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature, 583, 699–710. - PMC - PubMed
-
- Boyle, A. P. , Hong, E. L. , Hariharan, M. , Cheng, Y. , Schaub, M. A. , Kasowski, M. , Karczewski, K. J. , Park, J. , Hitz, B. C. , Weng, S. , Cherry, J. M. , & Snyder, M. (2012). Annotation of functional variation in personal genomes using RegulomeDB. Genome Research, 22, 1790–1797. - PMC - PubMed
-
- Caraceni, A. , Hanks, G. , Kaasa, S. , Bennett, M. I. , Brunelli, C. , Cherny, N. , Dale, O. , De Conno, F. , Fallon, M. , Hanna, M. , Haugen, D. F. , Juhl, G. , King, S. , Klepstad, P. , Laugsand, E. A. , Maltoni, M. , Mercadante, S. , Nabal, M. , Pigni, A. , … European Palliative Care Research Collaborative (EPCRC), European Association for Palliative Care (EAPC) . (2012). Use of opioid analgesics in the treatment of cancer pain: Evidence‐based recommendations from the EAPC. The Lancet Oncology, 13, e58–e68. - PubMed

