Genomic epidemiology and genetic characteristics of clinical Campylobacter species cocirculating in West Bengal, India, 2019, using whole genome analysis
- PMID: 39629976
- PMCID: PMC11784092
- DOI: 10.1128/aac.01108-24
Genomic epidemiology and genetic characteristics of clinical Campylobacter species cocirculating in West Bengal, India, 2019, using whole genome analysis
Abstract
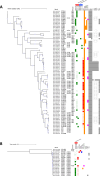
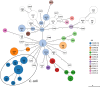
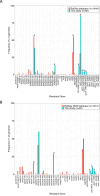
Campylobacter species are the most common pathogens responsible for foodborne gastroenteritis worldwide. India is a region with frequent diarrheal infections and a high level of Campylobacter infection incidence, but the detailed genomic information is limited. This study aimed to characterize 112 isolates of Campylobacter from diarrhea patients at two hospitals in Kolkata, West Bengal, by whole genome analysis. The Campylobacter isolates consisted of 90 C. jejuni, 20 C. coli, and 2 C. lari isolates. Multilocus sequence typing analysis revealed that the largest sequence type (ST) populations were ST-2131 in C. jejuni and ST-830 in C. coli and seven novel STs were found in C. jejuni and one in C. coli. Notably, ST-2131, which is rarely seen elsewhere, was positive for a sialylated LOS-related gene (wlaN +neuA + cstIII) associated with Guillain-Barré syndrome. Antibiotic resistance factors predicted from the genome sequence included blaOXA variants (58.9%), tet(O) (54.5%), tet(W) (0.9%), ant(6)-Ia (0.9%), mutation in GyrA (T86I, T86I+D90N, T86I+P104S, T86I+D90N+P104S) (79.5%), and mutation in 23S rRNA (A2075G) (12.5%). In addition to the high drug resistance of Campylobacter in Kolkata, Campylobacter pathogens were circulating that may be associated with Guillain-Barré syndrome. This study indicates the importance of genomic analysis in the surveillance of pathogens, which provides genomic information on genetic diversity, virulence mechanisms, and determinants of antimicrobial resistance.
Keywords: Campylobacter jejuni/coli; phylogenetic analysis; plasmid structure; whole genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921 - DOI - PMC - PubMed
-
- Lertpiriyapong K, Gamazon ER, Feng Y, Park DS, Pang J, Botka G, Graffam ME, Ge Z, Fox JG. 2012. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One 7:e42842. doi: 10.1371/journal.pone.0042842 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

