Targeting Caveolin-1 in Multiple Myeloma Cells Enhances Chemotherapy and Natural Killer Cell-Mediated Immunotherapy
- PMID: 39630017
- PMCID: PMC11789597
- DOI: 10.1002/advs.202408373
Targeting Caveolin-1 in Multiple Myeloma Cells Enhances Chemotherapy and Natural Killer Cell-Mediated Immunotherapy
Abstract
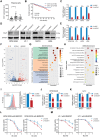
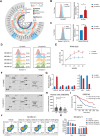
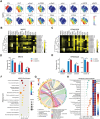
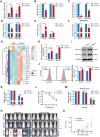
The cell membrane transport capacity and surface targets of multiple myeloma (MM) cells heavily influence chemotherapy and immunotherapy. Here, it is found that caveolin-1 (CAV1), a primary component of membrane lipid rafts and caveolae, is highly expressed in MM cells and is associated with MM progression and drug resistance. CAV1 knockdown decreases MM cell adhesion to stromal cells and attenuates cell adhesion-mediated drug resistance to bortezomib. CAV1 inhibition in MM cells enhances natural killer cell-mediated cytotoxicity through increasing CXCL10, SLAMF7, and CD112. CAV1 suppression reduces mitochondrial membrane potential, increases reactive oxygen species, and inhibits autophagosome-lysosome fusion, resulting in the disruption of redox homeostasis. Additionally, CAV1 knockdown enhances glutamine addiction by increasing ASCT2 and LAT1 and dysregulates glutathione metabolism. As a result of CAV1 inhibition, MM cells are more sensitive to starvation, glutamine depletion, and glutamine transporter inhibition, and grow more slowly in vivo in a mouse model treated with bortezomib. The observation that CAV1 inhibition modulated by 6-mercaptopurine, daidzin, and statins enhances the efficacy of bortezomib in vitro and in vivo highlights the translational significance of these FDA-approved drugs in improving MM outcomes. These data demonstrate that CAV1 serves as a potent therapeutic target for enhancing chemotherapy and immunotherapy for MM.
Keywords: caveolin‐1; glutathione metabolism; immunotherapy; multiple myeloma; natural killer cell; redox homeostasis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- a) Moreau P., Hulin C., Perrot A., Arnulf B., Belhadj K., Benboubker L., Bene M. C., Zweegman S., Caillon H., Caillot D., Corre J., Delforge M., Dejoie T., Doyen C., Facon T., Sonntag C., Fontan J., Mohty M., Jie K. S., Karlin L., Kuhnowski F., Lambert J., Leleu X., Macro M., Orsini‐Piocelle F., Roussel M., Stoppa A. M., van de Donk N., Wuilleme S., Broijl A., et al., Lancet Oncol. 2021, 22, 1378; - PubMed
- b) Mateos M. V., Cavo M., Blade J., Dimopoulos M. A., Suzuki K., Jakubowiak A., Knop S., Doyen C., Lucio P., Nagy Z., Pour L., Cook M., Grosicki S., Crepaldi A., Liberati A. M., Campbell P., Shelekhova T., Yoon S. S., Iosava G., Fujisaki T., Garg M., Krevvata M., Chen Y., Wang J., Kudva A., Ukropec J., Wroblewski S., Qi M., Kobos R., San‐Miguel J., Lancet 2020, 395, 132; - PubMed
- c) Usmani S. Z., Quach H., Mateos M. V., Landgren O., Leleu X., Siegel D., Weisel K., Gavriatopoulou M., Oriol A., Rabin N., Nooka A., Qi M., Beksac M., Jakubowiak A., Ding B., Zahlten‐Kumeli A., Yusuf A., Dimopoulos M., Lancet Oncol. 2022, 23, 65. - PubMed
-
- a) Manier S., Salem K. Z., Park J., Landau D. A., Getz G., Ghobrial I. M., Nat. Rev. Clin. Oncol. 2017, 14, 100; - PubMed
- b) Merz M., Merz A. M. A., Wang J., Wei L., Hu Q., Hutson N., Rondeau C., Celotto K., Belal A., Alberico R., Block A. W., Mohammadpour H., Wallace P. K., Tario J., Luce J., Glenn S. T., Singh P., Herr M. M., Hahn T., Samur M., Munshi N., Liu S., McCarthy P. L., Hillengass J., Nat. Commun. 2022, 13, 807; - PMC - PubMed
- c) Zhang H., Du Z., Tu C., Zhou X., Menu E., Wang J., Cancer Res. 2023, 84, 39. - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFC2304205/National Key Research and Development Program of China
- 82100224/National Natural Science Foundation of China
- 2024A1515013250/Guangdong Basic and Applied Basic Research Foundation
- 202235397/Tertiary Education Scientific Research Project of Guangzhou Municipal Education Bureau
- 2024ZDZX2054/Projects in Key Areas of General Universities in Guangdong Province
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
