BRAFV600E induces key features of LCH in iPSCs with cell type-specific phenotypes and drug responses
- PMID: 39630039
- PMCID: PMC11867135
- DOI: 10.1182/blood.2024026066
BRAFV600E induces key features of LCH in iPSCs with cell type-specific phenotypes and drug responses
Abstract
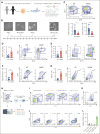
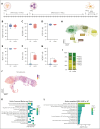
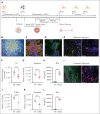
Langerhans cell histiocytosis (LCH) is a clonal hematopoietic disorder defined by tumorous lesions containing CD1a+/CD207+ cells. Two severe complications of LCH are systemic hyperinflammation and progressive neurodegeneration. The scarcity of primary samples and lack of appropriate models limit our mechanistic understanding of LCH pathogenesis and affect patient care. We generated a human in vitro model for LCH using induced pluripotent stem cells (iPSCs) harboring the BRAFV600E mutation, the most common genetic driver of LCH. We show that BRAFV600E/WT iPSCs display myelomonocytic skewing during hematopoiesis and spontaneously differentiate into CD1a+/CD207+ cells that are similar to lesional LCH cells and are derived from a CD14+ progenitor. We show that BRAFV600E modulates the expression of key transcription factors regulating monocytic differentiation and leads to an upregulation of proinflammatory molecules and LCH marker genes early during myeloid differentiation. In vitro drug testing revealed that BRAFV600E-induced transcriptomic changes are reverted upon treatment with mitogen-activated protein kinase (MAPK) pathway inhibitors (MAPKis). Importantly, MAPKis do not affect myeloid progenitors but reduce only the mature CD14+ cell population. Furthermore, iPSC-derived neurons (iNeurons) cocultured with BRAFV600E/WT iPSC-derived microglia-like cells, differentiated from iPSC-derived CD34+ progenitors, exhibit signs of neurodegeneration with neuronal damage and release of neurofilament light chain. In summary, the iPSC-based model described here provides a platform to investigate the effects of BRAFV600E in different hematopoietic cell types and provides a tool to compare and identify novel approaches for the treatment of BRAFV600E-driven diseases.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




Comment in
-
Deciphering Langerhans cell histiocytosis.Blood. 2025 Feb 20;145(8):790-791. doi: 10.1182/blood.2024027473. Blood. 2025. PMID: 39976946 No abstract available.
References
-
- Cantwell-Dorris ER, O'Leary JJ, Sheils OM. BRAFV600E: implications for carcinogenesis and molecular therapy. Mol Cancer Ther. 2011;10(3):385–394. - PubMed
-
- Yu RC, Chu C, Buluwela L, Chu AC. Clonal proliferation of Langerhans cells in Langerhans cell histiocytosis. Lancet. 1994;343(8900):767–768. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

