A trafficking regulatory subnetwork governs αVβ6 integrin-HER2 cross-talk to control breast cancer invasion and drug resistance
- PMID: 39630893
- PMCID: PMC11616693
- DOI: 10.1126/sciadv.adk9944
A trafficking regulatory subnetwork governs αVβ6 integrin-HER2 cross-talk to control breast cancer invasion and drug resistance
Abstract
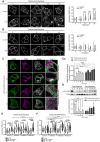
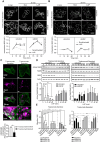
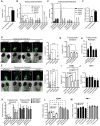
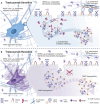
HER2 and αVβ6 integrin are independent predictors of breast cancer survival and metastasis. We identify an αVβ6/HER2 cross-talk mechanism driving invasion, which is dysregulated in drug-resistant HER2+ breast cancer cells. Proteomic analyses reveal ligand-bound αVβ6 recruits HER2 and a trafficking subnetwork, comprising guanosine triphosphatases RAB5 and RAB7A and the Rab regulator guanine nucleotide dissociation inhibitor 2 (GDI2). The RAB5/RAB7A/GDI2 functional module mediates direct cross-talk between αVβ6 and HER2, affecting receptor trafficking and signaling. Acute exposure to trastuzumab increases recruitment of the subnetwork to αVβ6, but trastuzumab resistance decouples GDI2 recruitment. GDI2, RAB5, and RAB7A cooperate to regulate migration and transforming growth factor-β activation to promote invasion. However, these mechanisms are dysregulated in trastuzumab-resistant cells. In patients, RAB5A, RAB7A, and GDI2 expression correlates with patient survival and αVβ6 expression predicts relapse following trastuzumab treatment. Thus, the RAB5/RAB7A/GDI2 subnetwork regulates αVβ6-HER2 cross-talk to drive breast cancer invasion but is subverted in trastuzumab-resistant cells to drive αVβ6-independent and HER2-independent tumor progression.
Figures




References
-
- Ross J. S., Fletcher J. A., The HER-2/neu oncogene in breast cancer: Prognostic factor, predictive factor, and target for therapy. Oncologist 3, 237–252 (1998). - PubMed
-
- Yarden Y., Sliwkowski M. X., Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2, 127–137 (2001). - PubMed
-
- Andersson J., Linderholm B., Greim G., Lindh B., Lindman H., Tennvall J., Tennvall-Nittby L., Pettersson-Skold D., Sverrisdottir A., Soderberg M., Klaar S., Bergh J., A population-based study on the first forty-eight breast cancer patients receiving trastuzumab (Herceptin) on a named patient basis in Sweden. Acta Oncol. 41, 276–281 (2002). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

