Genetic and microenvironmental evolution of colorectal liver metastases under chemotherapy
- PMID: 39631402
- PMCID: PMC11722126
- DOI: 10.1016/j.xcrm.2024.101838
Genetic and microenvironmental evolution of colorectal liver metastases under chemotherapy
Abstract
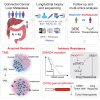
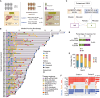
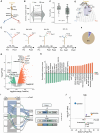
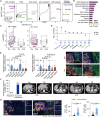
Drug resistance limits the efficacy of chemotherapy for colorectal cancer liver metastasis (CRLM). However, the evolution of CRLM during drug treatment remains poorly elucidated. Multi-omics and treatment response data from 115 samples of 49 patients with CRLM undergoing bevacizumab (BVZ)-based chemotherapy show little difference in genomic alterations in 92% of cases, while remarkable differences are observed at the transcriptomic level. By decoupling intrinsic and acquired resistance, we find that hepatocyte and myeloid cell infiltration contribute to 38.5% and 23.1% of acquired resistance, respectively. Importantly, SMAD4 mutations and chr20q copy-number gain are associated with intrinsic chemoresistance. Gene interference experiments suggest that SMAD4R361H/C mutations confer BVZ and 5-fluorouracil (5-FU) resistance through STAT3 signaling. Notably, supplementing BVZ and 5-FU with the STAT3 inhibitor GB201 restores therapeutic efficacy in SMAD4R361H/C cancer cells. Our study uncovers the evolutionary dynamics of CRLM and its microenvironment during treatment and offers strategies to overcome drug resistance.
Keywords: Bevacizumab; SMAD4 mutation; cancer genomics; chemoresistance; chr20q copy-number gain; colorectal liver metastases; tumor microenvironment.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Hurwitz H., Fehrenbacher L., Novotny W., Cartwright T., Hainsworth J., Heim W., Berlin J., Baron A., Griffing S., Holmgren E., et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004;350:2335–2342. - PubMed
-
- Cremolini C., Loupakis F., Antoniotti C., Lupi C., Sensi E., Lonardi S., Mezi S., Tomasello G., Ronzoni M., Zaniboni A., et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306–1315. - PubMed
-
- Bennouna J., Sastre J., Arnold D., Österlund P., Greil R., Van Cutsem E., von Moos R., Viéitez J.M., Bouché O., Borg C., et al. Continuation of bevacizumab after first progression in metastatic colorectal cancer (ML18147): a randomised phase 3 trial. Lancet Oncol. 2013;14:29–37. - PubMed
-
- Masi G., Vivaldi C., Fornaro L., Lonardi S., Buccianti P., Sainato A., Marcucci L., Martignetti A., Luca Urso E.D., Castagna M., et al. Total neoadjuvant approach with FOLFOXIRI plus bevacizumab followed by chemoradiotherapy plus bevacizumab in locally advanced rectal cancer: the TRUST trial. Eur. J. Cancer. 2019;110:32–41. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

