Integral Solvent-Induced Protein Precipitation for Target-Engagement Studies in Plasmodium falciparum
- PMID: 39631773
- PMCID: PMC11650652
- DOI: 10.1021/acsinfecdis.4c00418
Integral Solvent-Induced Protein Precipitation for Target-Engagement Studies in Plasmodium falciparum
Abstract
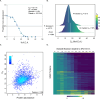
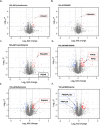
The limited understanding of the mechanism of action (MoA) of several antimalarials and the rise of drug resistance toward existing malaria therapies emphasizes the need for new strategies to uncover the molecular target of compounds in Plasmodium falciparum. Integral solvent-induced protein precipitation (iSPP) is a quantitative mass spectrometry-based (LC-MS/MS) proteomics technique. The iSPP leverages the change in solvent-induced denaturation of the drug-bound protein relative to its unbound state, allowing identification of the direct drug-protein target without the need to modify the drug. Here, we demonstrate proof-of-concept of iSPP in P. falciparum (Pf) lysate. At first, we profiled the solvent-induced denaturation behavior of the Pf proteome, generating denaturation curves and determining the melting concentration (CM) of 2712 proteins. We then assessed the extent of stabilization of three antimalarial target proteins in multiple organic solvent gradients, allowing for a rational selection of an optimal solvent gradient. Subsequently, we validated iSPP by successfully showing target-engagement of several standard antimalarials. The iSPP assay allows the testing of multiple conditions within reasonable LC-MS/MS measurement time. Furthermore, it requires a minimal amount of protein input, reducing culturing time and simplifying protein extraction. We envision that iSPP will be useful as a complementary tool for MoA studies for next-generation antimalarials.
Keywords: antimalarial; drug discovery; iSPP; proteomics; solvents; target-engagement.
Conflict of interest statement
The authors declare the following competing financial interest(s): H.H. is a co-founder and shareholder of OmicScouts GmbH, a proteomics and chemical company.
Figures



References
-
- World Health Organization . World Malaria Report; World Health Organization, 2023.
-
- Kamala T.; Van H. N.; Anna R.-U.; Quang P. B.; Do Manh H.; Evi P.; Pieter G.; Van V. N.; Thanh D. T.; Alfred A.-N.; Umberto D.; Annette E. Delayed Parasite Clearance after Treatment with Dihydroartemisinin-Piperaquine in Plasmodium Falciparum Malaria Patients in Central Vietnam. Antimicrob. Agents Chemother. 2014, 58 (12), 7049–7055. 10.1128/aac.02746-14. - DOI - PMC - PubMed
-
- Van der Pluijm R. W.; Tripura R.; Hoglund R. M.; Pyae Phyo A.; Lek D.; ul Islam A.; Anvikar A. R.; Satpathi P.; Satpathi S.; Behera P. K.; Tripura A.; Baidya S.; Onyamboko M.; Chau N. H.; Sovann Y.; Suon S.; Sreng S.; Mao S.; Oun S.; Yen S.; Amaratunga C.; Chutasmit K.; Saelow C.; Runcharern R.; Kaewmok W.; Hoa N. T.; Thanh N. V.; Hanboonkunupakarn B.; Callery J. J.; Mohanty A. K.; Heaton J.; Thant M.; Gantait K.; Ghosh T.; Amato R.; Pearson R. D.; Jacob C. G.; Gonçalves S.; Mukaka M.; Waithira N.; Woodrow C. J.; Grobusch M. P.; van Vugt M.; Fairhurst R. M.; Cheah P. Y.; Peto T. J.; von Seidlein L.; Dhorda M.; Maude R. J.; Winterberg M.; Thuy-Nhien N. T.; Kwiatkowski D. P.; Imwong M.; Jittamala P.; Lin K.; Hlaing T. M.; Chotivanich K.; Huy R.; Fanello C.; Ashley E.; Mayxay M.; Newton P. N.; Hien T. T.; Valecha N.; Smithuis F.; Pukrittayakamee S.; Faiz A.; Miotto O.; Tarning J.; Day N. P. J.; White N. J.; Dondorp A. M.; van der Pluijm R. W.; Tripura R.; Hoglund R. M.; Phyo A. P.; Lek D.; ul Islam A.; Anvikar A. R.; Satpathi P.; Satpathi S.; Behera P. K.; Tripura A.; Baidya S.; Onyamboko M.; Chau N. H.; Sovann Y.; Suon S.; Sreng S.; Mao S.; Oun S.; Yen S.; Amaratunga C.; Chutasmit K.; Saelow C.; Runcharern R.; Kaewmok W.; Hoa N. T.; Thanh N. V.; Hanboonkunupakarn B.; Callery J. J.; Mohanty A. K.; Heaton J.; Thant M.; Gantait K.; Ghosh T.; Amato R.; Pearson R. D.; Jacob C. G.; Gonçalves S.; Mukaka M.; Waithira N.; Woodrow C. J.; Grobusch M. P.; van Vugt M.; Fairhurst R. M.; Cheah P. Y.; Peto T. J.; von Seidlein L.; Dhorda M.; Maude R. J.; Winterberg M.; Thuy-Nhien N. T.; Kwiatkowski D. P.; Imwong M.; Jittamala P.; Lin K.; Hlaing T. M.; Chotivanich K.; Huy R.; Fanello C.; Ashley E.; Mayxay M.; Newton P. N.; Hien T. T.; Valeche N.; Smithuis F.; Pukrittayakamee S.; Faiz A.; Miotto O.; Tarning J.; Day N. P. J.; White N. J.; Dondorp A. M. Triple Artemisinin-Based Combination Therapies versus Artemisinin-Based Combination Therapies for Uncomplicated Plasmodium Falciparum Malaria: A Multicentre, Open-Label, Randomised Clinical Trial. Lancet 2020, 395 (10233), 1345–1360. 10.1016/S0140-6736(20)30552-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

