In-Source Collision-Induced Dissociation (CID) Improves Higher-Energy Collisional Dissociation (HCD)-Dependent Fragmentation of ADP-Ribosyl Peptides
- PMID: 39632390
- PMCID: PMC11617611
- DOI: 10.1002/rcm.9961
In-Source Collision-Induced Dissociation (CID) Improves Higher-Energy Collisional Dissociation (HCD)-Dependent Fragmentation of ADP-Ribosyl Peptides
Abstract
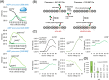
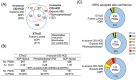
Rationale: ADP-ribosylation is a posttranslational modification whose higher-energy collisional dissociation (HCD) products are dominated by complete or partial modification losses, complicating peptide sequencing and acceptor site localization efforts. We tested whether in-source collision-induced dissociation (CID) performed on a quadrupole-Orbitrap could convert ADPr to the smaller phosphoribose-H2O derivative to facilitate HCD-dependent peptide sequencing.
Methods: ADP-ribosyl (ADPr) peptides derived from the human macrophage-like cell line THP-1 were analyzed on a quadrupole-Orbitrap. We monitored the dissociation of ADPr (+ 541.061 Da) to phosphoribosyl-H2O (+ 193.997 Da) peptides while varying the source and high-field asymmetric waveform ion mobility mass spectrometry (FAIMS) compensation voltages. Xcorr and ptmRS were used to evaluate peptide sequencing and acceptor site confidence, respectively. Phosphoribosyl-H2O acceptor sites were compared with those determined by electron-transfer higher-energy collision dissociation (EThcD), performed on a quadrupole-ion trap-Orbitrap.
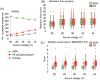
Results: In-source CID of ADPr peptides to their phosphoribosyl-H2O derivatives increased with increasing source voltage (up to 50 V), as judged by monitoring the corresponding modification loss ([adenosine monophosphate/AMP]+) and the number of identified phosphoribosyl-H2O peptide identifications. The average Xcorr increased from 1.36 (ADPr) to 2.26 (phosphoribosyl-H2O), similar to that achieved with EThcD for ADPr peptides (2.29). The number of high-confidence acceptor sites (> 95%) also increased, from 31% (ADPr) to 70% (phosphoribosyl-H2O), which was comparable to EThcD (70%).
Conclusions: In-source CID converts ADP-ribosyl to phosphoribosyl-H2O peptides that are more amenable to HCD-dependent peptide sequencing, providing an alternative method for acceptor site determination when ETD-based methods are not available.
Keywords: ADP‐ribosylation; electron transfer dissociation; in‐source collision‐induced dissociation; ribosylome.
© 2024 The Author(s). Rapid Communications in Mass Spectrometry published by John Wiley & Sons Ltd.
Conflict of interest statement
TK and YN are employees of Kowa Company, Ltd., Nagoya, Japan, but also current visiting scientists at Brigham and Women's Hospital and Harvard Medical School when the study was conducted.
Figures




Similar articles
-
Combining Higher-Energy Collision Dissociation and Electron-Transfer/Higher-Energy Collision Dissociation Fragmentation in a Product-Dependent Manner Confidently Assigns Proteomewide ADP-Ribose Acceptor Sites.Anal Chem. 2017 Feb 7;89(3):1523-1530. doi: 10.1021/acs.analchem.6b03365. Epub 2017 Jan 13. Anal Chem. 2017. PMID: 28035797
-
A Study into the ADP-Ribosylome of IFN-γ-Stimulated THP-1 Human Macrophage-like Cells Identifies ARTD8/PARP14 and ARTD9/PARP9 ADP-Ribosylation.J Proteome Res. 2019 Apr 5;18(4):1607-1622. doi: 10.1021/acs.jproteome.8b00895. Epub 2019 Mar 21. J Proteome Res. 2019. PMID: 30848916 Free PMC article.
-
A Novel Spectral Annotation Strategy Streamlines Reporting of Mono-ADP-ribosylated Peptides Derived from Mouse Liver and Spleen in Response to IFN-γ.Mol Cell Proteomics. 2022 Apr;21(4):100153. doi: 10.1016/j.mcpro.2021.100153. Epub 2021 Sep 28. Mol Cell Proteomics. 2022. PMID: 34592425 Free PMC article.
-
Mono-ADP-Ribosylation of Peptides: An Overview of Synthetic and Chemoenzymatic Methodologies.Chembiochem. 2024 Dec 16;25(24):e202400440. doi: 10.1002/cbic.202400440. Epub 2024 Sep 5. Chembiochem. 2024. PMID: 38984757 Free PMC article. Review.
-
Identification and analysis of ADP-ribosylated proteins.Curr Top Microbiol Immunol. 2015;384:33-50. doi: 10.1007/82_2014_424. Curr Top Microbiol Immunol. 2015. PMID: 25113886 Review.
References
-
- Honjo T., Nishizuka Y., and Hayaishi O., “Diphtheria Toxin‐Dependent Adenosine Diphosphate Ribosylation of Aminoacyl Transferase II and Inhibition of Protein Synthesis,” Journal of Biological Chemistry 243, no. 12 (1968): 3553–3555. - PubMed
-
- Schreiber V., Dantzer F., Ame J. C., and de Murcia G., “Poly (ADP‐Ribose): Novel Functions for an Old Molecule,” Nature Reviews. Molecular Cell Biology 7, no. 7 (2006): 517–528. - PubMed
-
- Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M. O., and Nielsen M. L., “Proteome‐Wide Identification of Poly (ADP‐Ribosyl)ation Targets in Different Genotoxic Stress Responses,” Molecular Cell 52, no. 2 (2013): 272–285. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
