Dynamics of the blood plasma proteome during hyperacute HIV-1 infection
- PMID: 39632834
- PMCID: PMC11618498
- DOI: 10.1038/s41467-024-54848-0
Dynamics of the blood plasma proteome during hyperacute HIV-1 infection
Abstract
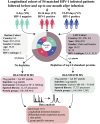
The complex dynamics of protein expression in plasma during hyperacute HIV-1 infection and its relation to acute retroviral syndrome, viral control, and disease progression are largely unknown. Here, we quantify 1293 blood plasma proteins from 157 longitudinally linked plasma samples collected before, during, and after hyperacute HIV-1 infection of 54 participants from four sub-Saharan African countries. Six distinct longitudinal expression profiles are identified, of which four demonstrate a consistent decrease in protein levels following HIV-1 infection. Proteins involved in inflammatory responses, immune regulation, and cell motility are significantly altered during the transition from pre-infection to one month post-infection. Specifically, decreased ZYX and SCGB1A1 levels, and increased LILRA3 levels are associated with increased risk of acute retroviral syndrome; increased NAPA and RAN levels, and decreased ITIH4 levels with viral control; and increased HPN, PRKCB, and ITGB3 levels with increased risk of disease progression. Overall, this study provides insight into early host responses in hyperacute HIV-1 infection, and present potential biomarkers and mechanisms linked to HIV-1 disease progression and viral load.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

