Kinesin-like motor protein KIF23 maintains neural stem and progenitor cell pools in the developing cortex
- PMID: 39632980
- PMCID: PMC11729872
- DOI: 10.1038/s44318-024-00327-7
Kinesin-like motor protein KIF23 maintains neural stem and progenitor cell pools in the developing cortex
Abstract
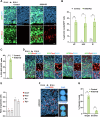
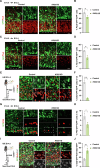
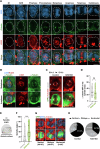
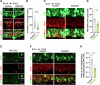
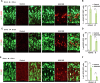
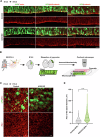
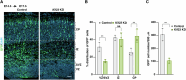
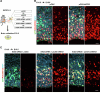
Accurate mitotic division of neural stem and progenitor cells (NSPCs) is crucial for the coordinated generation of progenitors and mature neurons, which determines cortical size and structure. While mutations in the kinesin-like motor protein KIF23 gene have been recently linked to microcephaly in humans, the underlying mechanisms remain elusive. Here, we explore the pivotal role of KIF23 in embryonic cortical development. We characterize the dynamic expression of KIF23 in the cortical NSPCs of mice, ferrets, and humans during embryonic neurogenesis. Knockdown of Kif23 in mice results in precocious neurogenesis and neuronal apoptosis, attributed to an accelerated cell cycle exit, likely resulting from disrupted mitotic spindle orientation and impaired cytokinesis. Additionally, KIF23 depletion perturbs the apical surface structure of NSPCs by affecting the localization of apical junction proteins. We further demonstrate that the phenotypes induced by Kif23 knockdown are rescued by introducing wild-type human KIF23, but not by a microcephaly-associated variant. Our findings unveil a previously unexplored role of KIF23 in neural stem and progenitor cell maintenance via regulating spindle orientation and apical structure in addition to cytokinesis, shedding light on microcephaly pathogenesis.
Keywords: Apoptosis; Cytokinesis; Kinesin Motor Proteins; Microcephaly; Neurogenesis.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

