Macrophages excite muscle spindles with glutamate to bolster locomotion
- PMID: 39633045
- PMCID: PMC11735391
- DOI: 10.1038/s41586-024-08272-5
Macrophages excite muscle spindles with glutamate to bolster locomotion
Erratum in
-
Author Correction: Macrophages excite muscle spindles with glutamate to bolster locomotion.Nature. 2025 Sep;645(8081):E4. doi: 10.1038/s41586-025-09550-6. Nature. 2025. PMID: 40841479 Free PMC article. No abstract available.
Abstract
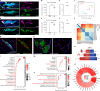
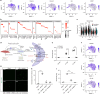
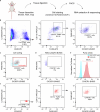
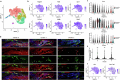
The stretch reflex is a fundamental component of the motor system that orchestrates the coordinated muscle contractions underlying movement. At the heart of this process lie the muscle spindles (MS), specialized receptors finely attuned to fluctuations in tension within intrafusal muscle fibres. The tension variation in the MS triggers a series of neuronal events including an initial depolarization of sensory type Ia afferents that subsequently causes the activation of motoneurons within the spinal cord1,2. This neuronal cascade culminates in the execution of muscle contraction, underscoring a presumed closed-loop mechanism between the musculoskeletal and nervous systems. By contrast, here we report the discovery of a new population of macrophages with exclusive molecular and functional signatures within the MS that express the machinery for synthesizing and releasing glutamate. Using mouse intersectional genetics with optogenetics and electrophysiology, we show that activation of MS macrophages (MSMP) drives proprioceptive sensory neuron firing on a millisecond timescale. MSMP activate spinal circuits, motor neurons and muscles by means of a glutamate-dependent mechanism that excites the MS. Furthermore, MSMP respond to neural and muscle activation by increasing the expression of glutaminase, enabling them to convert the uptaken glutamine released by myocytes during muscle contraction into glutamate. Selective silencing or depletion of MSMP in hindlimb muscles disrupted the modulation of the stretch reflex for force generation and sensory feedback correction, impairing locomotor strategies in mice. Our results have identified a new cellular component, the MSMP, that directly regulates neural activity and muscle contraction. The glutamate-mediated signalling of MSMP and their dynamic response to sensory cues introduce a new dimension to our understanding of sensation and motor action, potentially offering innovative therapeutic approaches in conditions that affect sensorimotor function.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











References
-
- Sherrington, C. S. in Scientific and Medical Knowledge Production, 1796–1918 (ed. Boddice, R.) 217–253 (Routledge, 2023).
-
- Liddell, E. G. T. & Sherrington, C. S. Reflexes in response to stretch (myotatic reflexes). Proc. R. Soc. Lond. Series B. Biol. Sci.96, 212–242 (1924).
-
- Creed, R. S., Denny-Brown, D., Eccles, J. C., Liddell, E. G. T. & Sherrington, C. S. Reflex Activity of the Spinal Cord (Clarendon Press, 1932).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

