RANK drives structured intestinal epithelial expansion during pregnancy
- PMID: 39633049
- PMCID: PMC11666467
- DOI: 10.1038/s41586-024-08284-1
RANK drives structured intestinal epithelial expansion during pregnancy
Abstract
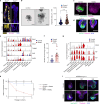
During reproduction, multiple species such as insects and all mammals undergo extensive physiological and morphological adaptions to ensure health and survival of the mother and optimal development of the offspring. Here we report that the intestinal epithelium undergoes expansion during pregnancy and lactation in mammals. This enlargement of the intestinal surface area results in a novel geometry of expanded villi. Receptor activator of nuclear factor-κΒ (RANK, encoded by TNFRSF11A) and its ligand RANKL were identified as a molecular pathway involved in this villous expansion of the small intestine in vivo in mice and in intestinal mouse and human organoids. Mechanistically, RANK-RANKL protects gut epithelial cells from cell death and controls the intestinal stem cell niche through BMP receptor signalling, resulting in the elongation of villi and a prominent increase in the intestinal surface. As a transgenerational consequence, babies born to female mice that lack Rank in the intestinal epithelium show reduced weight and develop glucose intolerance after metabolic stress. Whereas gut epithelial remodelling in pregnancy/lactation is reversible, constitutive expression of an active form of RANK is sufficient to drive intestinal expansion followed by loss of villi and stem cells, and prevents the formation of Apcmin-driven small intestinal stem cell tumours. These data identify RANK-RANKL as a pathway that drives intestinal epithelial expansion in pregnancy/lactation, one of the most elusive and fundamental tissue remodelling events in mammalian life history and evolution.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


















References
-
- Arck, P. C. & Hecher, K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat. Med.19, 548–556 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

