Fetal hepatocytes protect the HSPC genome via fetuin-A
- PMID: 39633051
- PMCID: PMC11711094
- DOI: 10.1038/s41586-024-08307-x
Fetal hepatocytes protect the HSPC genome via fetuin-A
Erratum in
-
Author Correction: Fetal hepatocytes protect the HSPC genome via fetuin-A.Nature. 2025 Mar;639(8053):E3. doi: 10.1038/s41586-025-08714-8. Nature. 2025. PMID: 39966683 Free PMC article. No abstract available.
Abstract
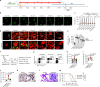
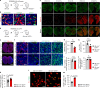
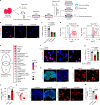
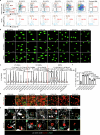
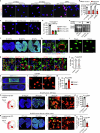
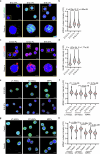
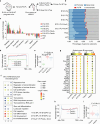
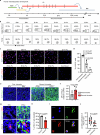
The maintenance of genomic integrity in rapidly proliferating cells is a substantial challenge during embryonic development1-3. Although numerous cell-intrinsic mechanisms have been revealed4-7, little is known about genome-protective effects and influences of developmental tissue microenvironments on tissue-forming cells. Here we show that fetal liver hepatocytes provide protection to haematopoietic stem and progenitor cell (HSPC) genomes. Lineage tracing and depletion in mice demonstrated that delayed hepatocyte development in early fetal livers increased the chromosomal instability of newly colonizing HSPCs. In addition, HSPCs developed tolerance to genotoxins in hepatocyte-conditioned medium, suggesting that hepatocytes protect the HSPC genome in a paracrine manner. Proteomic analyses demonstrated the enrichment of fetuin-A in hepatocyte-conditioned medium but not in early fetal livers. Fetuin-A activates a Toll-like receptor pathway to prevent pathogenic R-loop accumulation in HSPCs undergoing DNA replication and gene transcription in the fetal liver. Numerous haematopoietic regulatory genes frequently involved in leukaemogenic mutations are associated with R-loop-enriched regions. In Fetua-knockout mice, HSPCs showed increased genome instability and susceptibility to malignancy induction. Moreover, low concentrations of fetuin-A correlated with the oncogenesis of childhood leukaemia. Therefore, we uncover a mechanism operating in developmental tissues that offers tissue-forming cell genome protection and is implicated in developmental-related diseases.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Gaillard, H. & Aguilera, A. Transcription as a threat to genome integrity. Annu. Rev. Biochem.85, 291–317 (2016). - PubMed
-
- Aguilera, A. & Garcia-Muse, T. Causes of genome instability. Annu. Rev. Genet.47, 1–32 (2013). - PubMed
-
- Hoeijmakers, J. H. Genome maintenance mechanisms for preventing cancer. Nature411, 366–374 (2001). - PubMed
-
- Branzei, D. & Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol.11, 208–219 (2010). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

