Rarγ-Foxa1 signaling promotes luminal identity in prostate progenitors and is disrupted in prostate cancer
- PMID: 39633177
- PMCID: PMC11772605
- DOI: 10.1038/s44319-024-00335-y
Rarγ-Foxa1 signaling promotes luminal identity in prostate progenitors and is disrupted in prostate cancer
Abstract
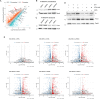
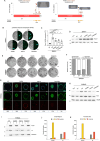
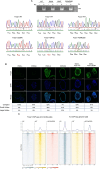
Retinoic acid (RA) signaling is a master regulator of vertebrate development with crucial roles in body axis orientation and tissue differentiation, including in the reproductive system. However, a mechanistic understanding of how RA signaling governs cell lineage identity is often missing. Here, leveraging prostate organoid technology, we show that RA signaling orchestrates the commitment of adult mouse prostate progenitors to glandular identity, epithelial barrier integrity, and specification of prostatic lumen. RA-dependent RARγ activation promotes the expression of Foxa1, which synergizes with the androgen pathway for luminal expansion, cytoarchitecture and function. FOXA1 mutations are common in prostate and breast cancers, though their pathogenic mechanism is incompletely understood. Combining functional genetics with structural modeling of FOXA1 folding and chromatin binding analyses, we discover that FOXA1F254E255 is a loss-of-function mutation compromising its transcriptional function and luminal fate commitment of prostate progenitors. Overall, we define RA as an instructive signal for glandular identity in adult prostate progenitors. Importantly, we identify cancer-associated FOXA1 indels affecting residue F254 as loss-of-function mutations promoting dedifferentiation of adult prostate progenitors.
Keywords: FOXA1; Organoids; Prostate; Retinoic Acid.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. FCh is a consultant for Dompè Pharmaceuticals SPA (not related to this work). All other authors declare no competing interests.
Figures








Update of
-
Rarγ -Foxa1 signaling promotes luminal identity in prostate progenitors and is disrupted in prostate cancer.bioRxiv [Preprint]. 2024 Mar 8:2024.03.06.583256. doi: 10.1101/2024.03.06.583256. bioRxiv. 2024. Update in: EMBO Rep. 2025 Jan;26(2):443-469. doi: 10.1038/s44319-024-00335-y. PMID: 38496627 Free PMC article. Updated. Preprint.
References
-
- Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, Zhang Z, Cho H, DiLoreto R, Chhangawala S, Liu Y et al (2019) Gene Expression Omnibus GSE128867. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128867 [DATASET]
-
- Alfthan O, Tarkkanen J, Gröhn P, Heinonen E, Pyrhönen S (1983) Tigason® (Etretinate) in prevention of recurrence of superficial bladder tumors. Eur Urol 9:6–9 - PubMed
MeSH terms
Substances
Grants and funding
- AIRC-IG 27893/Fondazione AIRC per la ricerca sul cancro ETS (AIRC)
- MFAG-20344/Fondazione AIRC per la ricerca sul cancro ETS (AIRC)
- CDA2014/Giovanni Armenise-Harvard Foundation (GAHF)
- PRIN 20174PLLYN/Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR)
- 2015-2024/Core funding from the Department CIBIO
- W81XWH-18-1-0424/U.S. Department of Defense (DOD)
- Core Facilities/EC | European Regional Development Fund (ERDF)
- PhD Fellowship/University of Trento
- Ateneo_Starting Grants Young Researchers_2019/University of Trento
- R01 CA238005/CA/NCI NIH HHS/United States
- 23-0321/Worldwide Cancer Research (WCR)
- FTRT/Fondazione Trentina per la Ricerca sui Tumori (FTRT)
- U01 CA261822/CA/NCI NIH HHS/United States
- FUV 2017/Fondazione Umberto Veronesi (Umberto Veronesi Foundation)
- MFAG 2017-ID 20621/Fondazione AIRC per la ricerca sul cancro ETS (AIRC)
- PhD Fellowship/Fondazione Pezcoller (Pezcoller Foundation)
- FUV 2016/Fondazione Umberto Veronesi (Umberto Veronesi Foundation)
LinkOut - more resources
Full Text Sources
Medical

