Unveiling the bacterial diversity and potential of the Avicennia marina ecosystem for enhancing plant resilience to saline conditions
- PMID: 39633419
- PMCID: PMC11619459
- DOI: 10.1186/s40793-024-00642-w
Unveiling the bacterial diversity and potential of the Avicennia marina ecosystem for enhancing plant resilience to saline conditions
Abstract
Background: Avicennia marina ecosystems are critical for coastal protection, water quality enhancement, and biodiversity support. These unique ecosystems thrive in extreme saline conditions and host a diverse microbiome that significantly contributes to plant resilience and growth. Global food security is increasingly threatened by crop yield losses due to abiotic stresses, including saline soils. Traditional plant breeding for salt tolerance is both costly and time-consuming. This study explores the potential of bacteria from A. marina to enhance plant growth under saline conditions, emphasizing their ecological significance.
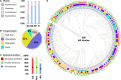
Results: We analyzed the microbiome of A. marina from the Red Sea coast using high-throughput Illumina sequencing and culture-dependent methods across various compartments (bulk soil, rhizosphere, rhizoplane, roots, and leaves). Our findings revealed distinct compartment-specific microbial communities, with Proteobacteria being the dominant phylum. Functional predictions indicated diverse microbial roles in metal uptake and plant growth promotion (PGP). Remarkably, our culture-dependent methods allowed us to recover 56% of the bacterial diversity present in the microbiome, resulting in the isolation and characterization of 256 bacterial strains. These isolates were screened for PGP traits, including salt and heat tolerance, siderophore production, and pectinase activity. Out of the 77 bacterial isolates tested, 11 demonstrated a significant ability to enhance Arabidopsis growth under salt stress.
Conclusions: Our study highlights the ecological significance of mangrove microbiomes and the potential of culture collections in offering innovative solutions for ecological restoration and crop production in saline conditions. The unique collection of mangrove bacteria, particularly from the rhizosphere and endophytes, showcases significant PGP traits and stress tolerance capabilities. These findings emphasize the importance of functional traits, such as salt tolerance, in the recruitment of endophytic bacteria by plants over taxonomic affiliation. The identified bacterial strains hold potential not only for developing biofertilizers to improve crop productivity but also for ecological restoration projects aimed at rehabilitating saline-degraded lands, thereby contributing to overall ecosystem health and sustainability.
Keywords: Avicennia marina; And Plant-microbial interaction; Biostimulants; Global food security; Halophilic bacteria; Salt stress tolerance.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Consent for publication: There is no conflict to consent for publication. Competing interests: The authors declare no competing interests. Conflict of interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Effects of Salinity on Assembly Characteristics and Function of Microbial Communities in the Phyllosphere and Rhizosphere of Salt-Tolerant Avicennia marina Mangrove Species.Microbiol Spectr. 2023 Feb 6;11(2):e0300022. doi: 10.1128/spectrum.03000-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36744884 Free PMC article.
-
Investigating the bacterial community of gray mangroves (Avicennia marina) in coastal areas of Tabuk region.PeerJ. 2024 Oct 18;12:e18282. doi: 10.7717/peerj.18282. eCollection 2024. PeerJ. 2024. PMID: 39434799 Free PMC article.
-
Diversity of Bacterial Microbiota of Coastal Halophyte Limonium sinense and Amelioration of Salinity Stress Damage by Symbiotic Plant Growth-Promoting Actinobacterium Glutamicibacter halophytocola KLBMP 5180.Appl Environ Microbiol. 2018 Sep 17;84(19):e01533-18. doi: 10.1128/AEM.01533-18. Print 2018 Oct 1. Appl Environ Microbiol. 2018. PMID: 30054358 Free PMC article.
-
Microbially Mediated Plant Salt Tolerance and Microbiome-based Solutions for Saline Agriculture.Biotechnol Adv. 2016 Nov 15;34(7):1245-1259. doi: 10.1016/j.biotechadv.2016.08.005. Epub 2016 Aug 30. Biotechnol Adv. 2016. PMID: 27587331 Review.
-
From swamp to field: how genes from mangroves and its associates can enhance crop salinity tolerance.Mol Biol Rep. 2024 Apr 29;51(1):598. doi: 10.1007/s11033-024-09539-w. Mol Biol Rep. 2024. PMID: 38683409 Review.
References
-
- United Nations DoEaSA, Population Division. World Population Prospects 2022: Summary of Results. UN DESA/POP/2022/TR/NO 3 2022.
-
- Qadir M, Quillérou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD. Economics of salt-induced land degradation and restoration. Nat Resour Forum. 2014;38(4):282–95.
LinkOut - more resources
Full Text Sources
Miscellaneous
