Forced MyD88 signaling in microglia impacts the production and survival of regenerated retinal neurons
- PMID: 39633708
- PMCID: PMC11614808
- DOI: 10.3389/fcell.2024.1495586
Forced MyD88 signaling in microglia impacts the production and survival of regenerated retinal neurons
Abstract
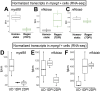
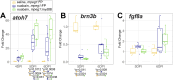
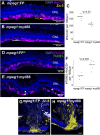
Inflammation and microglia appear to be key factors influencing the outcome of retinal regeneration following acute retinal damage. Despite such findings, direct connection of microglia-specific inflammatory factors as drivers of regenerative responses in the retina are still not defined, and intracellular pathways activated to stimulate such signals from microglia are currently unknown. We became interested in MyD88 regulation in microglia because transcriptomic datasets suggest myd88 could be regulated temporally in zebrafish microglia responding to damage in the central nervous system. MyD88 is an intracellular molecular adaptor that initiates signaling cascades downstream of several innate immune receptors, and probably most well-known for inducing gene expression of pro-inflammatory factors. Using zebrafish, which spontaneously regenerate retinal neurons after acute retinal damage, we studied the effects of overactivation of MyD88 signaling in microglia and macrophages on the Müller glia-mediated regenerative response. Our results indicate that increased MyD88 signaling in microglia/macrophages impacts the initial response of Müller glia entering a regenerative response after acute, neurotoxin-induced retinal damage to inner retinal neurons. In addition, increased MyD88 signaling in microglia/macrophages resulted in reduced survival of inner retinal neurons in regenerated retinas. This work supports the idea that temporal control of inflammatory signaling is a key component in the production of MG-derived progenitors yet further indicates that such control is important for differentiation and survival of regenerated neurons.
Keywords: MyD88; Müller glia; NFkB; inflammation; microglia; regeneration; retina; zebrafish.
Copyright © 2024 Rumford, Grieshaber, Lewiston, Reed, Long and Mitchell.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

