Proteomics and Incident Kidney Failure in Individuals With CKD: The African American Study of Kidney Disease and Hypertension and the Boston Kidney Biopsy Cohort
- PMID: 39634331
- PMCID: PMC11615895
- DOI: 10.1016/j.xkme.2024.100921
Proteomics and Incident Kidney Failure in Individuals With CKD: The African American Study of Kidney Disease and Hypertension and the Boston Kidney Biopsy Cohort
Abstract
Rationale & objective: Individuals with chronic kidney disease (CKD) are at increased risk of morbidity and mortality, particularly as they progress to kidney failure. Identifying circulating proteins that underlie kidney failure development may guide the discovery of new targets for intervention.
Study design: Prospective cohort.
Setting & participants: 703 African American Study of Kidney Disease and Hypertension (AASK) and 434 Boston Kidney Biopsy Cohort (BKBC) participants with baseline proteomics data.
Exposures: Circulating proteins measured using SomaScan.
Outcomes: Kidney failure, defined as dialysis initiation or kidney transplantation.
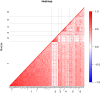
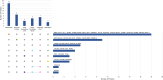
Analytical approach: Using adjusted Cox models, we studied associations of 6,284 circulating proteins with kidney failure risk separately in AASK and BKBC and meta-analyzed results. We then performed gene set enrichment analyses to identify underlying perturbations in biological pathways. In separate data sets with kidney-tissue level gene expression, we ascertained dominant regions of expression and correlated kidney tubular gene expression with fibrosis and estimated glomerular filtration rate (eGFR).
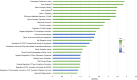
Results: Over median follow-up periods of 8.8 and 3.1 years, 210 AASK (mean age: 55 years, 39% female, mean GFR: 46 mL/min/1.73 m2) and 115 BKBC (mean age: 54 years, 47% female, mean eGFR: 51 mL/min/1.73 m2) participants developed kidney failure, respectively. We identified 143 proteins that were associated with incident kidney failure, of which only 1 (Testican-2) had a lower risk. Notable proteins included those related to vascular permeability (endothelial cell-selective adhesion molecule), glomerulosclerosis (ephrin-A1), glomerular development (ephrin-B2), intracellular sorting/transport (vesicular integral-membrane protein VIP36), podocyte effacement (pigment epithelium-derived factor), complement activation (complement decay-accelerating factor), and fibrosis (ephrin-A1, ephrin-B2, and pigment epithelium-derived factor). Gene set enrichment analyses detected overrepresented pathways that could be related to CKD progression, such as ephrin signaling, cell-cell junctions, intracellular transport, immune response, cell proliferation, and apoptosis. At the kidney level, glomerular expression predominated for genes corresponding to circulating proteins of interest, and several gene expression levels were correlated with eGFR and/or fibrosis.
Limitations: Possible residual confounding.
Conclusions: Multimodal data identified proteins and pathways associated with the development of kidney failure.
Keywords: ESAM; Proteomics; SPOCK2; ephrin; kidney failure.
Plain language summary
Circulating proteins that underlie the development of kidney failure may be new targets for treatment. In the current study of adults with chronic kidney disease, we evaluated over 6,000 proteins detected in blood and found more than 100 proteins whose levels were associated with new-onset kidney failure. Further investigation using gene expression data showed that the genes encoding these proteins were expressed in the kidney and involved in pathways of immune responses as well as cell signaling, structure, transport, and survival.
Figures

References
-
- United States Renal Data System . National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: 2021. 2021 USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

