Clustering of TP53 variants into functional classes correlates with cancer risk and identifies different phenotypes of Li-Fraumeni syndrome
- PMID: 39634561
- PMCID: PMC11615613
- DOI: 10.1016/j.isci.2024.111296
Clustering of TP53 variants into functional classes correlates with cancer risk and identifies different phenotypes of Li-Fraumeni syndrome
Abstract
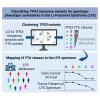
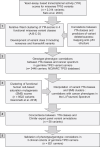
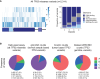
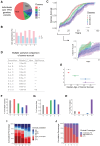
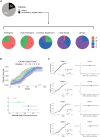
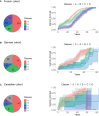
Li-Fraumeni syndrome (LFS) is a heterogeneous predisposition to an individually variable spectrum of cancers caused by pathogenic TP53 germline variants. We used a clustering method to assign TP53 missense variants to classes based on their functional activities in experimental assays assessing biological p53 functions. Correlations with LFS phenotypes were analyzed using the public germline TP53 mutation database and validated in three LFS clinical cohorts. Class A carriers recapitulated all phenotypic traits of fully penetrant LFS, whereas class B carriers showed a slightly less penetrant form dominated by specific cancers, consistent with the notion that these classes identify variants with distinct functional properties. Class C displayed a lower lifetime cancer risk associated with attenuated LFS features, consistent with the notion that these variants have hypomorphic features. Class D carriers showed low lifetime cancer risks inconsistent with LFS definitions. This classification of TP53 variants provides insights into structural/functional features causing pathogenicity.
Keywords: Genomics; Phenotyping.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Li F.P., Fraumeni J.F., Mulvihill J.J., Blattner W.A., Dreyfus M.G., Tucker M.A., Miller R.W. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48:5358–5362. - PubMed
-
- Kratz C.P., Freycon C., Maxwell K.N., Nichols K.E., Schiffman J.D., Evans D.G., Achatz M.I., Savage S.A., Weitzel J.N., Garber J.E., et al. Analysis of the Li-Fraumeni Spectrum Based on an International Germline TP53 Variant Data Set: An International Agency for Research on Cancer TP53 Database Analysis. JAMA Oncol. 2021;7:1800–1805. doi: 10.1001/jamaoncol.2021.4398. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

