Systems Neuroscience Computing in Python (SyNCoPy): a python package for large-scale analysis of electrophysiological data
- PMID: 39635648
- PMCID: PMC11614769
- DOI: 10.3389/fninf.2024.1448161
Systems Neuroscience Computing in Python (SyNCoPy): a python package for large-scale analysis of electrophysiological data
Abstract
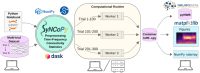
We introduce an open-source Python package for the analysis of large-scale electrophysiological data, named SyNCoPy, which stands for Systems Neuroscience Computing in Python. The package includes signal processing analyses across time (e.g., time-lock analysis), frequency (e.g., power spectrum), and connectivity (e.g., coherence) domains. It enables user-friendly data analysis on both laptop-based and high-performance computing systems. SyNCoPy is designed to facilitate trial-parallel workflows (parallel processing of trials), making it an ideal tool for large-scale analysis of electrophysiological data. Based on parallel processing of trials, the software can support very large-scale datasets via innovative out-of-core computation techniques. It also provides seamless interoperability with other standard software packages through a range of file format importers and exporters and open file formats. The naming of the user functions closely follows the well-established FieldTrip framework, which is an open-source MATLAB toolbox for advanced analysis of electrophysiological data.
Keywords: Granger causality spectra; big data; coherence spectra; electroencephalography (EEG); local field potential (LFP); magnetoencephalography (MEG); power spectra; spike train.
Copyright © 2024 Mönke, Schäfer, Parto-Dezfouli, Kajal, Fürtinger, Schmiedt and Fries.
Conflict of interest statement
PF has a patent on thin-film electrodes and is a member of the Advisory Board of CorTec GmbH (Freiburg, Germany). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Denker M., Köhler C., Jurkus R., Kern M., Kurth A. C., Kleinjohann A., et al. (2023). Elephant 0.13.0. Zenodo. doi: 10.5281/zenodo.8144467 - DOI
LinkOut - more resources
Full Text Sources

