A conserved cell-pole determinant organizes proper polar flagellum formation
- PMID: 39636223
- PMCID: PMC11620751
- DOI: 10.7554/eLife.93004
A conserved cell-pole determinant organizes proper polar flagellum formation
Abstract
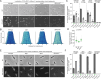
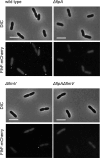
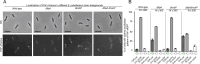
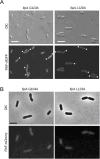
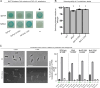
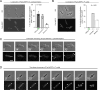
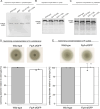
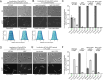
The coordination of cell cycle progression and flagellar synthesis is a complex process in motile bacteria. In γ-proteobacteria, the localization of the flagellum to the cell pole is mediated by the SRP-type GTPase FlhF. However, the mechanism of action of FlhF, and its relationship with the cell pole landmark protein HubP remain unclear. In this study, we discovered a novel protein called FipA that is required for normal FlhF activity and function in polar flagellar synthesis. We demonstrated that membrane-localized FipA interacts with FlhF and is required for normal flagellar synthesis in Vibrio parahaemolyticus, Pseudomonas putida, and Shewanella putrefaciens, and it does so independently of the polar localization mediated by HubP. FipA exhibits a dynamic localization pattern and is present at the designated pole before flagellar synthesis begins, suggesting its role in licensing flagellar formation. This discovery provides insight into a new pathway for regulating flagellum synthesis and coordinating cellular organization in bacteria that rely on polar flagellation and FlhF-dependent localization.
Keywords: Pseudomonas putida; cell biology; infectious disease; microbiology; shewanella putrefaciens; vibrio parahaemolyticus.
© 2024, Arroyo-Pérez, Hook et al.
Conflict of interest statement
EA, JH, AA, SW, TG, KT, SR No competing interests declared
Figures











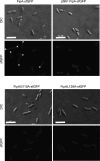
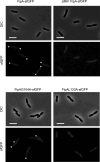
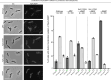
Update of
- doi: 10.1101/2023.09.20.558563
- doi: 10.7554/eLife.93004.1
- doi: 10.7554/eLife.93004.2
References
-
- Blagotinsek V, Schwan M, Steinchen W, Mrusek D, Hook JC, Rossmann F, Freibert SA, Kratzat H, Murat G, Kressler D, Beckmann R, Beeby M, Thormann KM, Bange G. An ATP-dependent partner switch links flagellar C-ring assembly with gene expression. PNAS. 2020;117:20826–20835. doi: 10.1073/pnas.2006470117. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

