Non-canonical CDK6 activity promotes cilia disassembly by suppressing axoneme polyglutamylation
- PMID: 39636239
- PMCID: PMC11619382
- DOI: 10.1083/jcb.202405170
Non-canonical CDK6 activity promotes cilia disassembly by suppressing axoneme polyglutamylation
Abstract
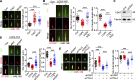
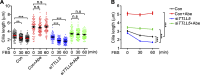
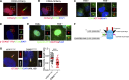
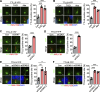
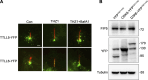
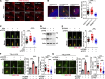
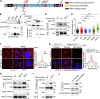
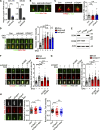
Tubulin polyglutamylation is a posttranslational modification that occurs primarily along the axoneme of cilia. Defective axoneme polyglutamylation impairs cilia function and has been correlated with ciliopathies, including Joubert Syndrome (JBTS). However, the precise mechanisms regulating proper axoneme polyglutamylation remain vague. Here, we show that cyclin-dependent kinase 6 (CDK6), but not its paralog CDK4, localizes to the cilia base and suppresses axoneme polyglutamylation by phosphorylating RAB11 family interacting protein 5 (FIP5) at site S641, a critical regulator of cilia import of glutamylases. S641 phosphorylation disrupts the ciliary recruitment of FIP5 and its association with RAB11, thereby reducing the ciliary import of glutamylases. Encouragingly, the FDA-approved CDK4/6 inhibitor Abemaciclib can effectively restore cilia function in JBTS cells with defective glutamylation. In summary, our study elucidates the regulatory mechanisms governing axoneme polyglutamylation and suggests that developing CDK6-specific inhibitors could be a promising therapeutic strategy to enhance cilia function in ciliopathy patients.
© 2024 He et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

