Human DBR1 deficiency impairs stress granule-dependent PKR antiviral immunity
- PMID: 39636299
- PMCID: PMC11619777
- DOI: 10.1084/jem.20240010
Human DBR1 deficiency impairs stress granule-dependent PKR antiviral immunity
Abstract
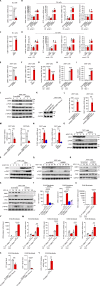
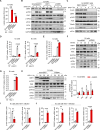
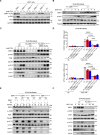
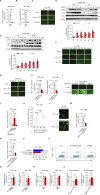
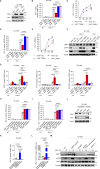
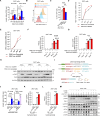
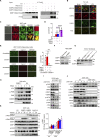
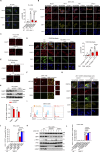
The molecular mechanism by which inborn errors of the human RNA lariat-debranching enzyme 1 (DBR1) underlie brainstem viral encephalitis is unknown. We show here that the accumulation of RNA lariats in human DBR1-deficient cells interferes with stress granule (SG) assembly, promoting the proteasome degradation of at least G3BP1 and G3BP2, two key components of SGs. In turn, impaired assembly of SGs, which normally recruit PKR, impairs PKR activation and activity against viruses, including HSV-1. Remarkably, the genetic ablation of PKR abolishes the corresponding antiviral effect of DBR1 in vitro. We also show that Dbr1Y17H/Y17H mice are susceptible to similar viral infections in vivo. Moreover, cells and brain samples from Dbr1Y17H/Y17H mice exhibit decreased G3BP1/2 expression and PKR phosphorylation. Thus, the debranching of RNA lariats by DBR1 permits G3BP1/2- and SG assembly-mediated PKR activation and cell-intrinsic antiviral immunity in mice and humans. DBR1-deficient patients are prone to viral disease because of intracellular lariat accumulation, which impairs G3BP1/2- and SG assembly-dependent PKR activation.
© 2024 Ru et al.
Conflict of interest statement
Disclosures: W. Fairbrother reported being a co-founder of Walah Scientific, a COVID breathalyzer company. W. Fairbrother is on the SAB of ReMIX. No other disclosures were reported.
Figures
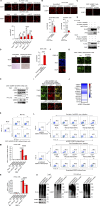



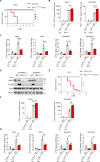
References
-
- Armakola, M., Higgins M.J., Figley M.D., Barmada S.J., Scarborough E.A., Diaz Z., Fang X., Shorter J., Krogan N.J., Finkbeiner S., et al. . 2012. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat. Genet. 44:1302–1309. 10.1038/ng.2434 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01AI088364/NH/NIH HHS/United States
- R01 NS072381/NS/NINDS NIH HHS/United States
- Rockefeller University
- 2023YFC2306100/National Key R&D Program of China
- UL1 TR001866/TR/NCATS NIH HHS/United States
- French Medical Research Foundation
- XDB0490000/Chinese Academy of Sciences
- HHMI/Howard Hughes Medical Institute/United States
- ANR-10-LABX-62-IBEID/French National Research Agency
- 31970855/National Natural Science Foundation of China
- R01 AI088364/AI/NIAID NIH HHS/United States
- Imagine Institute
- St. Giles Foundation
- General Atlantic Foundation
- Paris Cité University
- Institut National de la Santé et de la Recherche Médicale
- University of Science and Technology of China
LinkOut - more resources
Full Text Sources
Miscellaneous

