Targeting the PREX2/RAC1/PI3Kβ Signaling Axis Confers Sensitivity to Clinically Relevant Therapeutic Approaches in Melanoma
- PMID: 39636745
- PMCID: PMC11831108
- DOI: 10.1158/0008-5472.CAN-23-2814
Targeting the PREX2/RAC1/PI3Kβ Signaling Axis Confers Sensitivity to Clinically Relevant Therapeutic Approaches in Melanoma
Abstract
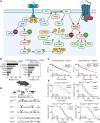
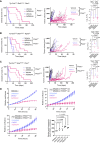
Metastatic melanoma remains a major clinical challenge. Large-scale genomic sequencing of melanoma has identified bona fide activating mutations in RAC1, which are associated with resistance to BRAF-targeting therapies. Targeting the RAC1-GTPase pathway, including the upstream activator PREX2 and the downstream effector PI3Kβ, could be a potential strategy for overcoming therapeutic resistance, limiting melanoma recurrence, and suppressing metastatic progression. Here, we used genetically engineered mouse models and patient-derived BRAFV600E-driven melanoma cell lines to dissect the role of PREX2 in melanomagenesis and response to therapy. Although PREX2 was dispensable for the initiation and progression of melanoma, its loss conferred sensitivity to clinically relevant therapeutics targeting the MAPK pathway. Importantly, genetic and pharmacologic targeting of PI3Kβ phenocopied PREX2 deficiency, sensitizing model systems to therapy. These data reveal a druggable PREX2/RAC1/PI3Kβ signaling axis in BRAF-mutant melanoma that could be exploited clinically. Significance: Cotargeting the MAPK and the PREX2/RAC1/PI3Kβ pathways has remarkable efficacy and outperforms monotherapy MAPK inhibition in BRAF-mutant melanoma, supporting the potential of this combination therapy for treating metastatic melanoma.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
C.A. Ford reports grants from Cancer Research UK and nonfinancial support from AstraZeneca during the conduct of the study. M. Foth reports that after completion of the work presented in this manuscript, she assumed a Research Scientist position at the Huntsman Cancer Institute, UT, where parts of her research project and salary are paid through a sponsored research agreement from Revolution Medicines to her current mentor, Dr. Martin McMahon. N. Sphyris reports a patent for inhibition of p38 MAPK for the treatment of cancer (Publication number: 20190125735; Filed: December 28, 2016; Publication date: May 2, 2019) pending. D.C. Hornigold reports being an employee and shareholder of AstraZeneca. J. Downward reports grants and personal fees from AstraZeneca and Vividion, grants from Bristol Myers Squibb and Novartis, and personal fees from Roche, Amgen, and ONO outside the submitted work. H.C.E. Welch reports grants from BBSRC during the conduct of the study. S.T. Barry reports being an employee and shareholder of AstraZeneca. O.J. Sansom reports grants from AstraZeneca during the conduct of the study, as well as grants from Novartis, AstraZeneca, Boehringer Ingelheim, and Cancer Research Horizons outside the submitted work. A.D. Campbell reports grants from Cancer Research UK and nonfinancial support from AstraZeneca PLC during the conduct of the study, as well as grants from AstraZeneca outside the submitted work, and reports funding from Novartis and Boehringer Ingelheim related to the development of novel therapeutic approaches in cancer. No disclosures were reported by the other authors.
Figures



References
-
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. . Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. - PubMed
-
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. . Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135–47. - PubMed
-
- Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res 2006;12:2301s–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

