Single-cell analysis reveals host S phase drives large T antigen expression during BK polyomavirus infection
- PMID: 39636788
- PMCID: PMC11620372
- DOI: 10.1371/journal.ppat.1012663
Single-cell analysis reveals host S phase drives large T antigen expression during BK polyomavirus infection
Abstract
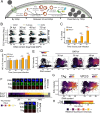
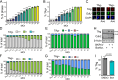
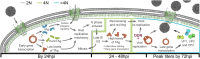
BK polyomavirus (BKPyV) is a major cause of kidney transplant failure, for which there are no antivirals. The current model is that BKPyV expresses TAg (large T antigen) early during infection, promoting cells to enter S phase where the viral DNA can access the host replication machinery. Here, we performed a single-cell analysis of viral TAg expression throughout the cell cycle to reveal that robust TAg expression required replication of the host DNA first. By using inhibitors that only affect host and not viral replication, we show that both TAg expression and viral production rely on an initial S phase. BKPyV is known to promote cellular re-replication, where the cell re-enters S phase from G2 phase (without passing through mitosis or G1 phase) to prolong S phase for viral replication. Thus, BKPyV infection results in cells with greater than 4N DNA content. We found that these subsequent rounds of replication of the host DNA relied on canonical host cell cycle machinery and regulators despite BKPyV infection. Together, these findings suggest a model for polyomavirus replication, where robust viral TAg expression depends on an initial host S phase and that BKPyV primarily replicates during host re-replication. Having a better understanding of the molecular events that are required for BKPyV production will help identify effective therapeutic targets against BKPyV.
Copyright: © 2024 Needham et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

