Orientia, Rickettsia, and the microbiome in rodent attached chiggers in North Carolina, USA
- PMID: 39637059
- PMCID: PMC11620566
- DOI: 10.1371/journal.pone.0311698
Orientia, Rickettsia, and the microbiome in rodent attached chiggers in North Carolina, USA
Abstract
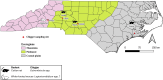
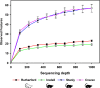
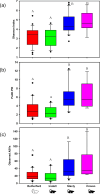
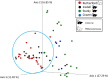
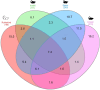
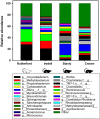
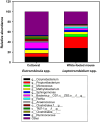
Chiggers are larval mites that pose a significant health risk globally via the spread of scrub typhus. However, fundamental studies into the bacterial microbiome in North America have never been considered. In this investigation, chiggers were collected in the wild from two locally common rodent host species (i.e., Sigmodon hispidus and Peromyscus leucopus) in three different ecoregions of North Carolina (NC), United States to investigate the composition of their bacterial communities, including potential pathogens. DNA was extracted from the chiggers, and the V3-V4 regions of the bacterial 16S rRNA gene were sequenced using next-generation sequencing (NGS). Alpha diversity metrics revealed significant differences in bacterial diversity among different collection counties. Beta diversity metrics also revealed that bacterial communities across counties were significantly different, suggesting changes in the microbiome as the environment changed. Specifically, we saw that the two western NC collection counties had similar bacterial composition as did the two eastern collection counties. In addition, we found that the chigger microbiome bacterial diversity and composition differed between rodent host species. The 16S rRNA sequence reads were assigned to 64 phyla, 106 orders, 199 families, and 359 genera. The major bacterial phylum was Actinobacteria. The most abundant species were in the genera Corynebacterium, Propionibacterium, class ZB2, and Methylobacterium. Sequences derived from potential pathogens within the genera Orientia and Rickettsia were also detected. Our findings provide the first insights into the ecology of chigger microbiomes in the US. Further research is required to determine if the potential pathogens found detected in chiggers are a threat to humans and wildlife.
Copyright: © 2024 Richardson et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

