Advantages of a genomic DNA-based next-generation sequencing assay for detection of mutant NPM1 measurable residual disease in AML
- PMID: 39637308
- PMCID: PMC11909430
- DOI: 10.1182/bloodadvances.2024014490
Advantages of a genomic DNA-based next-generation sequencing assay for detection of mutant NPM1 measurable residual disease in AML
Abstract
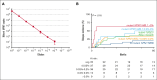
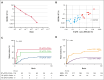
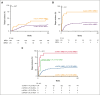
Mutations in the nucleophosmin-1 (NPM1) gene are among the most common molecular aberrations in acute myeloid leukemia (AML). Various studies have established mutant NPM1 (mNPM1) as a faithful molecular measurable residual disease (MRD) marker with prognostic significance. Assessment of prognostic mNPM1 is included in the European LeukemiaNet recommendations on MRD detection in AML. Because of recent advancements of promising drugs targeting mNPM1 AML, monitoring of mNPM1 MRD has gained interest, and is generally done by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR). However, these RT-qPCR assays use complementary DNA (cDNA) as input, are based on gene expression levels of mNPM1, and are generally limited to specific mNPM1 gene variants. The main advantages of next-generation sequencing (NGS) using genomic DNA as input are stability, independence of gene expression levels, and the ability to detect any NPM1 variant in a single assay. Here, we comprehensively investigated the applicability of NGS on DNA to detect mNPM1 MRD in a cohort of 119 (cDNA) and 310 (DNA) patients with mNPM1 AML in complete remission after 2 cycles of induction chemotherapy. We demonstrate high correlations in levels and prognostic value between RT-qPCR/cDNA and NGS/DNA approaches, postulating NGS/DNA as an attractive alternative to RT-qPCR. We report that the 2% mNPM1/ABL1 threshold by RT-qPCR/cDNA corresponds to an NGS/DNA variant allele frequency of 0.01%. The NGS/DNA threshold of >0.01% after 2 cycles of induction chemotherapy identifies significantly more patients with AML with an increased relapse risk than current RT-qPCR/cDNA assays. The prognostic significance of mNPM1 MRD appears greatest in patients with AML with FLT3-internal tandem duplications.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures
References
-
- Heath EM, Chan SM, Minden MD, Murphy T, Shlush LI, Schimmer AD. Biological and clinical consequences of NPM1 mutations in AML. Leukemia. 2017;31(4):798–807. - PubMed
-
- Dohner H, Wei AH, Appelbaum FR, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140(12):1345–1377. - PubMed
-
- Walter RB, Ofran Y, Wierzbowska A, et al. Measurable residual disease as a biomarker in acute myeloid leukemia: theoretical and practical considerations. Leukemia. 2021;35(6):1529–1538. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

