Modulation of TCR stimulation and pifithrin-α improve the genomic safety profile of CRISPR-engineered human T cells
- PMID: 39637860
- PMCID: PMC11722128
- DOI: 10.1016/j.xcrm.2024.101846
Modulation of TCR stimulation and pifithrin-α improve the genomic safety profile of CRISPR-engineered human T cells
Abstract
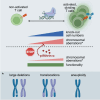
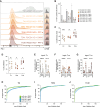
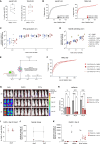
CRISPR-engineered chimeric antigen receptor (CAR) T cells are at the forefront of novel cancer treatments. However, several reports describe the occurrence of CRISPR-induced chromosomal aberrations. So far, measures to increase the genomic safety of T cell products focused mainly on the components of the CRISPR-Cas9 system and less on T cell-intrinsic features, such as their massive expansion after T cell receptor (TCR) stimulation. Here, we describe driving forces of indel formation in primary human T cells. Increased T cell activation and proliferation speed correlate with larger deletions. Editing of non-activated T cells reduces the risk of large deletions with the downside of reduced knockout efficiencies. Alternatively, the addition of the small-molecule pifithrin-α limits large deletions, chromosomal translocations, and aneuploidy in a p53-independent manner while maintaining the functionality of CRISPR-engineered T cells, including CAR T cells. Controlling T cell activation and pifithrin-α treatment are easily implementable strategies to improve the genomic integrity of CRISPR-engineered T cells.
Keywords: CAR T cell therapy; CRISPR engineering; T cell activation; aneuploidy; chromosomal aberrations; genomic integrity; human T cells; large deletions; pifithrin-alpha; translocations.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Ottaviano G., Georgiadis C., Gkazi S.A., Syed F., Zhan H., Etuk A., Preece R., Chu J., Kubat A., Adams S., et al. Phase 1 clinical trial of CRISPR-engineered CAR19 universal T cells for treatment of children with refractory B cell leukemia. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abq3010. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

