RuvBL1/2 reduce toxic dipeptide repeat protein burden in multiple models of C9orf72-ALS/FTD
- PMID: 39638345
- PMCID: PMC11629685
- DOI: 10.26508/lsa.202402757
RuvBL1/2 reduce toxic dipeptide repeat protein burden in multiple models of C9orf72-ALS/FTD
Abstract
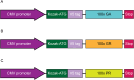
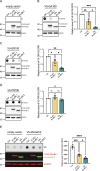
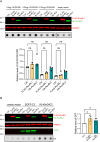
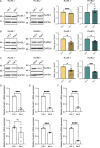
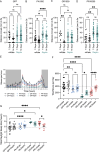
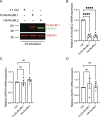
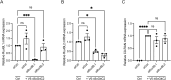
A G4C2 hexanucleotide repeat expansion in C9orf72 is the most common cause of amyotrophic lateral sclerosis and frontotemporal dementia (C9ALS/FTD). Bidirectional transcription and subsequent repeat-associated non-AUG (RAN) translation of sense and antisense transcripts leads to the formation of five dipeptide repeat (DPR) proteins. These DPRs are toxic in a wide range of cell and animal models. Therefore, decreasing RAN-DPRs may be of therapeutic benefit in the context of C9ALS/FTD. In this study, we found that C9ALS/FTD patients have reduced expression of the AAA+ family members RuvBL1 and RuvBL2, which have both been implicated in aggregate clearance. We report that overexpression of RuvBL1, but to a greater extent RuvBL2, reduced C9orf72-associated DPRs in a range of in vitro systems including cell lines, primary neurons from the C9-500 transgenic mouse model, and patient-derived iPSC motor neurons. In vivo, we further demonstrated that RuvBL2 overexpression and consequent DPR reduction in our Drosophila model was sufficient to rescue a number of DPR-related motor phenotypes. Thus, modulating RuvBL levels to reduce DPRs may be of therapeutic potential in C9ALS/FTD.
© 2024 Webster et al.
Conflict of interest statement
CP Webster and M Azzouz are coinventors on patents filled in the USA (US20230038479) and Europe (EP4061933) for the use of gene therapy vectors containing RuvBL1 and/or RuvBL2 “…for the treatment of neurodegenerative diseases that result from expression of polymorphic repeat expansions of GGGGCC in the first intron of the
Figures







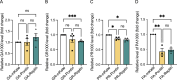



References
-
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, et al. (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646. 10.1016/j.neuron.2013.02.004 - DOI - PMC - PubMed
-
- Atanasio A, Decman V, White D, Ramos M, Ikiz B, Lee H-C, Siao C-J, Brydges S, LaRosa E, Bai Y, et al. (2016) C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep 6: 23204. 10.1038/srep23204 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
