Identification of cepharanthine as an effective inhibitor of African swine fever virus replication
- PMID: 39638605
- PMCID: PMC11622385
- DOI: 10.1080/22221751.2024.2429624
Identification of cepharanthine as an effective inhibitor of African swine fever virus replication
Abstract
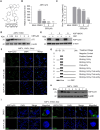
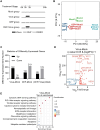
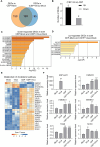
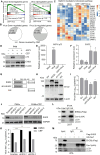
African swine fever virus (ASFV) causes highly contagious swine disease, African swine fever (ASF), thereby posing a severe socioeconomic threat to the global pig industry and underscoring that effective antiviral therapies are urgently required. To identify safe and efficient anti-ASFV compounds, a natural compound library was screened by performing an established cell-based ELISA in an ASFV-infected porcine alveolar macrophage (PAM) model. In total, 6 effective anti-ASFV compounds with low cytotoxicity were identified. Cepharanthine (CEP), a bisbenzylisoquinoline alkaloid, was the most potent inhibitor effect with an IC50 of 0.3223 μM. To further investigate the mechanism through which CEP inhibits ASFV replication, transcriptome profiles were generated in PAMs treated with CEP and/or infected with ASFV. ASFV infection dramatically altered immune response-associated gene expression. CEP treatment upregulated the expression of cholesterol biosynthesis-related genes, regardless of infection status. According to time-of-addition experiments, CEP primarily exerts its antiviral effect during the early stages of ASFV infection, specifically by inhibiting viral entry. Transcriptomic analysis suggested that CEP blocks ASFV entry through the clathrin-mediated endocytosis pathway by increasing EHD2 gene expression in macrophages. Disrupting EHD2 with small interfering RNA promoted ASFV entry into clathrin-positive vesicles. Finally, the protective effect of CEP in vivo was evaluated using ASFV-infected pigs. CEP could provide partial protection against ASFV infection, as indicated by an increase in survival time from 9.67 days to 16.67 days. Our findings imply that CEP exhibits potential antiviral activity against ASFV infection in PAMs, positioning it as a promising therapeutic strategy for ASF.
Keywords: African swine fever virus; EHD2; cepharanthine; cholesterol biosynthesis; transcriptome analysis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


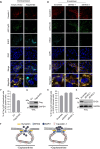
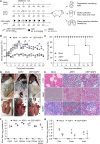
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
