Synaptic connectome of the Drosophila circadian clock
- PMID: 39638801
- PMCID: PMC11621569
- DOI: 10.1038/s41467-024-54694-0
Synaptic connectome of the Drosophila circadian clock
Abstract
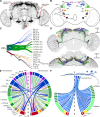
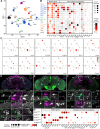
The circadian clock and its output pathways play a pivotal role in optimizing daily processes. To obtain insights into how diverse rhythmic physiology and behaviors are orchestrated, we have generated a comprehensive connectivity map of an animal circadian clock using the Drosophila FlyWire brain connectome. Intriguingly, we identified additional dorsal clock neurons, thus showing that the Drosophila circadian network contains ~240 instead of 150 neurons. We revealed extensive contralateral synaptic connectivity within the network and discovered novel indirect light input pathways to the clock neurons. We also elucidated pathways via which the clock modulates descending neurons that are known to regulate feeding and reproductive behaviors. Interestingly, we observed sparse monosynaptic connectivity between clock neurons and downstream higher-order brain centers and neurosecretory cells known to regulate behavior and physiology. Therefore, we integrated single-cell transcriptomics and receptor mapping to decipher putative paracrine peptidergic signaling by clock neurons. Our analyses identified additional novel neuropeptides expressed in clock neurons and suggest that peptidergic signaling significantly enriches interconnectivity within the clock network.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

