SARS-CoV-2 N protein recruits G3BP to double membrane vesicles to promote translation of viral mRNAs
- PMID: 39638802
- PMCID: PMC11621422
- DOI: 10.1038/s41467-024-54996-3
SARS-CoV-2 N protein recruits G3BP to double membrane vesicles to promote translation of viral mRNAs
Abstract
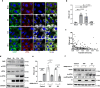
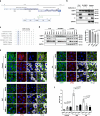
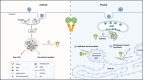
Ras-GTPase-activating protein SH3-domain-binding proteins (G3BP) are critical for the formation of stress granules (SGs) through their RNA- and ribosome-binding properties. SARS-CoV-2 nucleocapsid (N) protein exhibits strong binding affinity for G3BP and inhibits infection-induced SG formation soon after infection. To study the impact of the G3BP-N interaction on viral replication and pathogenesis in detail, we generated a mutant SARS-CoV-2 (RATA) that specifically lacks the G3BP-binding motif in the N protein. RATA triggers a stronger and more persistent SG response in infected cells, showing reduced replication across various cell lines, and greatly reduced pathogenesis in K18-hACE2 transgenic mice. At early times of infection, G3BP and WT N protein strongly colocalise with dsRNA and with non-structural protein 3 (nsp3), a component of the pore complex in double membrane vesicles (DMVs) from which nascent viral RNA emerges. Furthermore, G3BP-N complexes promote highly localized translation of viral mRNAs in the immediate vicinity of the DMVs and thus contribute to efficient viral gene expression and replication. In contrast, G3BP is absent from the DMVs in cells infected with RATA and translation of viral mRNAs is less efficient. This work provides a fuller understanding of the multifunctional roles of G3BP in SARS-CoV-2 infection.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery.PLoS Pathog. 2019 Jun 14;15(6):e1007842. doi: 10.1371/journal.ppat.1007842. eCollection 2019 Jun. PLoS Pathog. 2019. PMID: 31199850 Free PMC article.
-
Intrinsic Factors Behind Long COVID: VI. Combined Impact of G3BPs and SARS-CoV-2 Nucleocapsid Protein on the Viral Persistence and Long COVID.J Cell Biochem. 2025 May;126(5):e70038. doi: 10.1002/jcb.70038. J Cell Biochem. 2025. PMID: 40415285 Review.
-
Role of Chikungunya nsP3 in Regulating G3BP1 Activity, Stress Granule Formation and Drug Efficacy.Arch Med Res. 2021 Jan;52(1):48-57. doi: 10.1016/j.arcmed.2020.10.002. Epub 2020 Oct 31. Arch Med Res. 2021. PMID: 33131924
-
Disruption of Molecular Interactions between the G3BP1 Stress Granule Host Protein and the Nucleocapsid (NTD-N) Protein Impedes SARS-CoV-2 Virus Replication.Biochemistry. 2025 Feb 18;64(4):823-840. doi: 10.1021/acs.biochem.4c00536. Epub 2024 Dec 21. Biochemistry. 2025. PMID: 39708056
-
Research Progress on the Structure and Function of G3BP.Front Immunol. 2021 Aug 30;12:718548. doi: 10.3389/fimmu.2021.718548. eCollection 2021. Front Immunol. 2021. PMID: 34526993 Free PMC article. Review.
Cited by
-
Stress granules: Guardians of cellular health and triggers of disease.Neural Regen Res. 2026 Feb 1;21(2):588-597. doi: 10.4103/NRR.NRR-D-24-01196. Epub 2025 Feb 24. Neural Regen Res. 2026. PMID: 39995077 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

