Cell competition drives bronchiolization and pulmonary fibrosis
- PMID: 39639058
- PMCID: PMC11621346
- DOI: 10.1038/s41467-024-54997-2
Cell competition drives bronchiolization and pulmonary fibrosis
Abstract
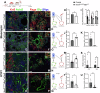
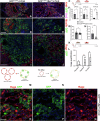
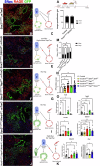
Idiopathic pulmonary fibrosis (IPF) is a progressive respiratory scarring disease arising from the maladaptive differentiation of lung stem cells into bronchial epithelial cells rather than into alveolar type 1 (AT1) cells, which are responsible for gas exchange. Here, we report that healthy lungs maintain their stem cells through tonic Hippo and β-catenin signaling, which promote Yap/Taz degradation and allow for low-level expression of the Wnt target gene Myc. Inactivation of upstream activators of the Hippo pathway in lung stem cells inhibits this tonic β-catenin signaling and Myc expression and promotes their Taz-mediated differentiation into AT1 cells. Vice versa, increased Myc in collaboration with Yap promotes the differentiation of lung stem cells along the basal and myoepithelial-like lineages allowing them to invade and bronchiolize the lung parenchyma in a process reminiscent of submucosal gland development. Our findings indicate that stem cells exhibiting the highest Myc levels become supercompetitors that drive remodeling, whereas loser cells with lower Myc levels terminally differentiate into AT1 cells.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






Update of
-
Cell competition drives bronchiolization and pulmonary fibrosis.Res Sq [Preprint]. 2024 Apr 22:rs.3.rs-4177351. doi: 10.21203/rs.3.rs-4177351/v1. Res Sq. 2024. Update in: Nat Commun. 2024 Dec 5;15(1):10624. doi: 10.1038/s41467-024-54997-2. PMID: 38746309 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 HL132156/HL/NHLBI NIH HHS/United States
- R01 HL126732/HL/NHLBI NIH HHS/United States
- T32 HL105355/HL/NHLBI NIH HHS/United States
- NIH R01 HL146461/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- NIH R01 HL132156/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

