AlzGenPred - CatBoost-based gene classifier for predicting Alzheimer's disease using high-throughput sequencing data
- PMID: 39639110
- PMCID: PMC11621786
- DOI: 10.1038/s41598-024-82208-x
AlzGenPred - CatBoost-based gene classifier for predicting Alzheimer's disease using high-throughput sequencing data
Abstract
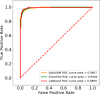
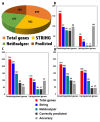
AD is a progressive neurodegenerative disorder characterized by memory loss. Due to the advancement in next-generation sequencing, an enormous amount of AD-associated genomics data is available. However, the information about the involvement of these genes in AD association is still a research topic. Therefore, AlzGenPred is developed to identify the AD-associated genes using machine-learning. A total of 13,504 features derived from eight sequence-encoding schemes were generated and evaluated using 16 machine learning algorithms. Network-based features significantly outperformed sequence-based features, effectively distinguishing AD-associated genes. In contrast, sequence-based features failed to classify accurately. To improve performance, we generated 24 fused features (6020 D) from sequence-based encodings, increasing accuracy by 5-7% using a two-step lightGBM-based recursive feature selection method. However, accuracy remained below 70% even after hyperparameter tuning. Therefore, network-based features were used to generate the CatBoost-based ML method AlzGenPred with 96.55% accuracy and 98.99% AUROC. The developed method is tested on the AlzGene dataset where it showed 96.43% accuracy. Then the model was validated using the transcriptomics dataset. AlzGenPred provides a reliable and user-friendly tool for identifying potential AD biomarkers, accelerating biomarker discovery, and advancing our understanding of AD. It is available at https://www.bioinfoindia.org/alzgenpred/ and https://github.com/shuklarohit815/AlzGenPred .
Keywords: Alzheimer’s disease; CatBoost; Machine learning; Network features; Neurofibrillary tangles.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures








References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

