Revisiting microgenderome: detecting and cataloguing sexually unique and enriched species in human microbiomes
- PMID: 39639265
- PMCID: PMC11622641
- DOI: 10.1186/s12915-024-02025-6
Revisiting microgenderome: detecting and cataloguing sexually unique and enriched species in human microbiomes
Abstract
Background: Microgenderome or arguably more accurately microsexome refers to studies on sexual dimorphism of human microbiomes aimed at investigating bidirectional interactions between human microbiomes, sex hormones, and immune systems. It is important because of its implications to disease susceptibility and therapy, in which men and women demonstrate divergence in many diseases especially autoimmune diseases. In a previous report [1], we presented analyses of several key ecological aspects of microgenderome by leveraging the large datasets of the HMP (human microbiome project) but failed to offer species-level composition differences such as sexually unique species (US) and enriched species (ES). Existing approaches, for such tasks, including differential species relative abundance analysis and differential network analysis, possess certain limitations given that virtually all rely on species abundance alone or are univariate, while ignoring species distribution information across samples. Obviously, it is both species abundance and distribution that shape/drive the structure and dynamics of human microbiomes, and both should be equally responsible for the universal heterogeneity of microbiomes including the sexual dimorphism.
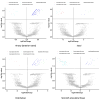
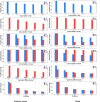
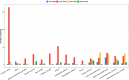
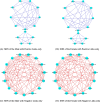
Results: Here, we fill the gap by taking advantages of a recently developed computational algorithm, species specificity, and specificity diversity (SSD) framework (refer to the companion article) to reanalyze the HMP and complementary seminovaginal microbiome datasets. The SSD framework can randomly search and catalogue the sexually specific unique/enriched species with statistical rigor, guided by species specificity (a synthetic metric of abundance and distribution) and specificity diversity (SD). The SSD framework reveals that men seem to have more unique species than women in their gut and reproductive system microbiomes, but women seem to have more unique species than men in the airway, oral, and skin microbiomes, which is likely due to sexual dimorphism in the hormone and immune systems. We further investigate co-dependency and heterogeneity of those sexually unique/enriched species across 15 body sites, with core/periphery network analyses.
Conclusions: This study not only produced sexually unique/enriched species in the human microbiomes and analyzed their codependency and heterogeneity but also further validated the robustness of the SSD framework presented in the companion article, by performing all negative control tests based on the HMP gut microbiome samples.
Keywords: Microgenderome; Permutation test; Sexually enriched species (ES); Sexually unique species (US); Species specificity (SS); Species specificity and specificity diversity (SSD) framework; Specificity diversity (SD).
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This is not applicable since the study does not involve any wet-lab experiments or survey on human or animal subjects, and all analyzed datasets are already available in public domain, as mentioned above. Consent for publication: All authors approved the submission for publication. Competing interests: The author declares no competing interests.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

