Feature Engineering for the Prediction of Scoliosis in 5q-Spinal Muscular Atrophy
- PMID: 39639589
- PMCID: PMC11670177
- DOI: 10.1002/jcsm.13599
Feature Engineering for the Prediction of Scoliosis in 5q-Spinal Muscular Atrophy
Abstract
Background: 5q-Spinal muscular atrophy (SMA) is now one of the 5% treatable rare diseases worldwide. As disease-modifying therapies alter disease progression and patient phenotypes, paediatricians and consulting disciplines face new unknowns in their treatment decisions. Conclusions made from historical patient data sets are now mostly limited, and new approaches are needed to ensure our continued best standard-of-care practices for this exceptional patient group. Here, we present a data-driven machine learning approach to a rare disease data set to predict spinal muscular atrophy (SMA)-associated scoliosis.
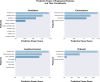
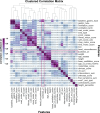
Methods: We collected data from 84 genetically confirmed 5q-SMA patients who have received novel SMA therapies. We performed expert domain knowledge-directed feature engineering, correlation and predictive power score (PPS) analyses for feature selection. To test the predictive performance of the selected features, we trained a Random Forest Classifier and evaluated model performance using standard metrics.
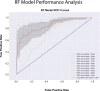
Results: The SMA data set consisted of 1304 visits and over 360 variables. We performed feature engineering for variables related to 'interventions', 'devices', 'orthosis', 'ventilation', 'muscle contractures' and 'motor milestones'. Through correlation and PPS analysis paired with expert domain knowledge feature selection, we identified relevant features for scoliosis prediction in SMA that included disease progression markers: Hammersmith Functional Motor Scale Expanded 'HFMSE' (PPS = 0.27) and 6-Minute Walk Test '6MWT' scores (PPS = 0.44), 'age' (PPS = 0.41) and 'weight' (PPS = 0.49), 'contractures' (PPS = 0.17), the use of 'assistive devices' (PPS = 0.39, 'ventilation' (PPS = 0.16) and the presence of 'gastric tubes' (PPS = 0.35) in SMA patients. These features were validated using expert domain knowledge and used to train a Random Forest Classifier with an observed accuracy of 0.82 and an average receiver operating characteristic (ROC) area of 0.87.
Conclusion: The introduction of disease-modifying SMA therapies, followed by the implementation of SMA in newborn screenings, has presented physicians with never-seen patients. We used feature engineering tools to overcome one of the main challenges when using data-driven approaches in rare disease data sets. Through predictive modelling of this data, we defined disease progression markers, which are easily assessed during patient visits and can help anticipate scoliosis onset. This highlights the importance of progressive features in the drug-induced revolution of this rare disease and further supports the ongoing efforts to update the SMA classification. We advocate for the consistent documentation of relevant progression markers, which will serve as a basis for data-driven models that physicians can use to update their best standard-of-care practices.
Keywords: feature engineering; gene therapy; machine learning; predictive power score; rare disease; spinal muscular atrophy.
© 2024 The Author(s). Journal of Cachexia, Sarcopenia and Muscle published by Wiley Periodicals LLC.
Conflict of interest statement
Claudia Weiß is on the honorary advisory board of Novartis, Roche and Biogen and has given an honorary presentation at conferences for Novartis. The other authors declare no conflicts of interest.
Figures




References
-
- Richter T., Nestler‐Parr S., Babela R., et al., “Rare Disease Terminology and Definitions—A Systematic Global Review: Report of the ISPOR Rare Disease Special Interest Group,” Value in Health 18 (2015): 906–914. - PubMed
-
- Wirth B., Karakaya M., Kye M. J., and Mendoza‐Ferreira N., “Twenty‐Five Years of Spinal Muscular Atrophy Research: From Phenotype to Genotype to Therapy, and What Comes Next,” Annual Review of Genomics and Human Genetics 21 (2020): 231–261. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

