CBP/p300 lysine acetyltransferases inhibit HIV-1 expression in latently infected T cells
- PMID: 39640574
- PMCID: PMC11617383
- DOI: 10.1016/j.isci.2024.111244
CBP/p300 lysine acetyltransferases inhibit HIV-1 expression in latently infected T cells
Abstract
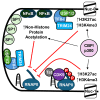
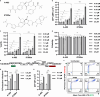
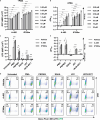
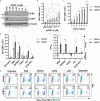
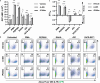
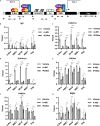
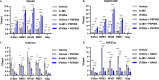
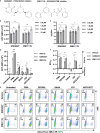
HIV-1 latency is regulated by chromatin modifying enzymes, and histone deacetylase inhibitors (HDACi) cause reactivation of provirus expression. Surprisingly, we observed that inhibitors of the CBP/p300 acetyltransferases also cause reversal of latency in T cells. CBP/p300 inhibitors synergize with various latency reversing agents to cause HIV-1 reactivation. In contrast, inhibition of CBP/p300 impaired reversal of latency by the HDACi SAHA, indicating that CBP/p300 must contribute to acetylation on the HIV-1 LTR associated with HDACi-mediated latency reversal. CBP/p300 inhibition caused loss of H3K27ac and H3K4me3 from the LTR, but did not affect association of the inhibitor protein BRD4. Furthermore, inhibition of the additional lysine acetyltransferases PCAF/GCN5 or KAT6A/KAT6B also caused reversal of latency, suggesting that protein acetylation has an inhibitory effect on HIV-1 expression. Collectively, these observations indicate that transcription from the HIV-1 LTR is controlled both positively and negatively by protein acetylation, likely including both histone and non-histone regulatory targets.
Keywords: Biological sciences; Immunology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Chun T.-W., Davey R.T., Ostrowski M., Shawn Justement J., Engel D., Mullins J.I., Fauci A.S. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 2000;6:757–761. doi: 10.1038/77481. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

