A scoping review of statistical methods to investigate colocalization between genetic associations and microRNA expression in osteoarthritis
- PMID: 39640910
- PMCID: PMC11617925
- DOI: 10.1016/j.ocarto.2024.100540
A scoping review of statistical methods to investigate colocalization between genetic associations and microRNA expression in osteoarthritis
Abstract
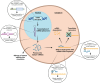
Background: Genetic colocalization analysis is a statistical method that evaluates whether two traits (e.g., osteoarthritis [OA] risk and microRNA [miRNA] expression levels) share the same or distinct genetic association signals in a locus typically identified in genome-wide association studies (GWAS). This method is useful for providing insights into the biological relevance of genetic association signals, particularly in intergenic regions, which can help to elucidate disease mechanisms in OA and other complex traits.
Objectives: To review the existing literature on genetic colocalization methods, assess their suitability for studying OA, and investigate their capacity to integrate miRNA data, while bearing in view their statistical assumptions.
Design: We followed scoping review methodology and used Covidence software for data management. Search terms for colocalization, GWAS, and genetic or statistical models were used in the databases MEDLINE and EMBASE, searched till March 4, 2024.
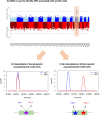
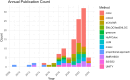
Results: Our search returned 546 peer-reviewed papers, of which 96 were included following title/abstract and full-text screening. Based on both cumulative and annual publication counts, the most cited method for colocalization analysis was coloc. Four papers examined OA-related phenotypes, and none examined miRNA. An approach to colocalization analysis using miRNA was postulated based on further hand-searching.
Conclusions: Colocalization analysis is a largely unexplored method in OA. Many of the approaches to colocalization analysis identified in this review, including the integration of GWAS and miRNA data, may help to elucidate genetic and epigenetic factors implicated in OA and other complex traits.
Keywords: Genetic architecture characterization; Genetic colocalization; Genetic pleiotropy; Genome-wide association study; MicroRNAs; Models; Statistical.
© 2024 The Author(s).
Conflict of interest statement
The authors have no relevant competing interests to declare.
Figures

References
Publication types
Associated data
LinkOut - more resources
Full Text Sources

