Genome-based newborn screening for severe childhood genetic diseases has high positive predictive value and sensitivity in a NICU pilot trial
- PMID: 39642868
- PMCID: PMC11639094
- DOI: 10.1016/j.ajhg.2024.10.020
Genome-based newborn screening for severe childhood genetic diseases has high positive predictive value and sensitivity in a NICU pilot trial
Abstract
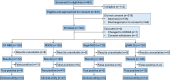
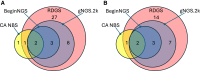
Large prospective clinical trials are underway or planned that examine the clinical utility and cost effectiveness of genome-based newborn screening (gNBS). One gNBS platform, BeginNGS, currently screens 53,575 variants for 412 severe childhood genetic diseases with 1,603 efficacious therapies. Retrospective evaluation of BeginNGS in 618,290 subjects suggests adequate sensitivity and positive predictive value (PPV) to proceed to prospective studies. To inform pivotal clinical trial design, we undertook a pilot clinical trial. We enrolled 120 infants in a regional neonatal intensive care unit (NICU) who were not under consideration for rapid diagnostic genome sequencing (RDGS). Each enrollee received BeginNGS and two index tests (California state NBS and RDGS). California NBS identified 4 of 4 true positive (TP) findings (TP rate 3.6%, sensitivity 100%) and 11 false positive (FP) findings (PPV 27%). RDGS identified 41 diagnostic findings in 36 neonates (diagnostic rate 30%). BeginNGS identified 5 of 6 on-target TP disorders (TP rate 4.2%, 95% confidence interval 1%-8%, sensitivity 83%) and no FPs (PPV 100%). Changes in management were anticipated following the return of 27 RDGS results in 25 enrollees (clinical utility [CU] 21%), 3 of 4 NBS TPs (CU 2.7%), and all BeginNGS TPs (CU 4.2%). The incidence of actionable genetic diseases in NICU infants not being considered for RDGS suggests (1) performance of RDGS in ∼20% of admissions misses many genetic diagnoses, (2) NICU enrollment in gNBS trials will greatly increase power to test endpoints, and (3) NICUs may be attractive for early implementation of consented BeginNGS screening.
Keywords: clinical trial; clinical utility; genome sequencing; infant; neonatal intensive care unit; newborn screening; positive predictive value; rapid diagnostic genome sequencing; sensitivity; severe childhood genetic disease.
Copyright © 2024 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.M.K. and S.S.M. are employees and shareholders of Illumina, Inc. W.R.M., Y.L., and T.D. are employees and shareholders of Alexion, AstraZeneca Rare Disease. J.L. is an employee and shareholder of TileDB, Inc.
Figures

References
-
- Collins F.S. Harper Collins; 2010. The Language of Life: DNA and the Revolution in Personalized Medicine; p. 208.
-
- Bhattacharjee A., Sokolsky T., Wyman S.K., Reese M.G., Puffenberger E., Strauss K., Morton H., Parad R.B., Naylor E.W. Development of DNA confirmatory and high-risk diagnostic testing for newborns using targeted next-generation DNA sequencing. Genet. Med. 2015;17:337–347. doi: 10.1038/gim.2014.117. - DOI - PubMed
-
- Bodian D.L., Klein E., Iyer R.K., Wong W.S.W., Kothiyal P., Stauffer D., Huddleston K.C., Gaither A.D., Remsburg I., Khromykh A., et al. Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1,696 neonates. Genet. Med. 2016;18:221–230. doi: 10.1038/gim.2015.111. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

