Macrophage Dvl2 deficiency promotes NOD1-Driven pyroptosis and exacerbates inflammatory liver injury
- PMID: 39644526
- PMCID: PMC11667066
- DOI: 10.1016/j.redox.2024.103455
Macrophage Dvl2 deficiency promotes NOD1-Driven pyroptosis and exacerbates inflammatory liver injury
Abstract
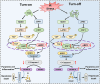
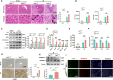
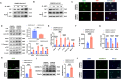
Dishevelled 2 (Dvl2) is a key mediator of the Wingless/Wnt signaling pathway that regulates cell proliferation, migration, and immune function. However, little is known about the role of macrophage Dvl2 in modulating NOD1-mediated pyroptosis and hepatocyte death in oxidative stress-induced inflammatory liver injury. In a mouse model of oxidative stress-induced liver inflammation, mice with myeloid-specific Dvl2 knockout (Dvl2M-KO) displayed exacerbated ischemia/reperfusion (IR) stress-induced hepatocellular damage with increased serum ALT levels, oxidative stress, and proinflammatory mediators. Unlike in Dvl2FL/FL controls, Dvl2M-KO enhanced NOD1, caspase-1, GSDMD, and NF-κB activation in liver macrophages after IR. Interestingly, IR stress enhanced YAP colocalized with HSF1 in Dvl2FL/FL macrophages, while macrophage Dvl2 deficiency reduced YAP and HSF1 colocalization in the nucleus under inflammatory conditions. Importantly, Dvl2 deletion diminished nuclear YAP interacted with HSF1 and augmented NOD1/caspase-1 and GSDMD activation in response to inflammatory stimulation. However, Dvl2 activation increased YAP interaction with HSF1 and activated HSF1 target gene eEF2, inhibiting NOD1/caspase-1, GSDMD, and NF-κB activity. Moreover, macrophage eEF2 deletion increased the NOD1-caspase-1 interaction, GSDMD activation, HMGB1 release, and hepatocyte LDH release after macrophage/hepatocyte co-culture. Adoptive transfer of eEF2-expressing macrophages in Dvl2M-KO mice alleviated IR-triggered liver inflammation and hepatocellular damage. Therefore, macrophage Dvl2 deficiency promotes NOD1-mediated pyroptosis and exacerbates IR-induced hepatocellular death by disrupting the YAP-HSF1 axis. eEF2 is crucial for modulating NOD1-driven pyroptosis, inflammatory response, and hepatocyte death. Our findings underscore a novel role of macrophage Dvl2 in modulating liver inflammatory injury and imply the therapeutic potential in organ IRI and transplant recipients.
Keywords: GSDMD; HSF1; TRPM7; YAP; eEF2.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

