CRISPR/Cas9-driven double modification of grapevine MLO6-7 imparts powdery mildew resistance, while editing of NPR3 augments powdery and downy mildew tolerance
- PMID: 39645650
- PMCID: PMC12034322
- DOI: 10.1111/tpj.17204
CRISPR/Cas9-driven double modification of grapevine MLO6-7 imparts powdery mildew resistance, while editing of NPR3 augments powdery and downy mildew tolerance
Abstract
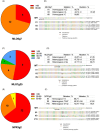
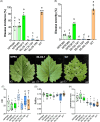
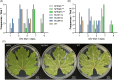
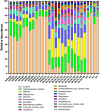
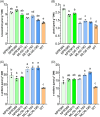
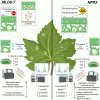
The implementation of genome editing strategies in grapevine is the easiest way to improve sustainability and resilience while preserving the original genotype. Among others, the Mildew Locus-O (MLO) genes have already been reported as good candidates to develop powdery mildew-immune plants. A never-explored grapevine target is NPR3, a negative regulator of the systemic acquired resistance. We report the exploitation of a cisgenic approach with the Cre-lox recombinase technology to generate grapevine-edited plants with the potential to be transgene-free while preserving their original genetic background. The characterization of three edited lines for each target demonstrated immunity development against Erysiphe necator in MLO6-7-edited plants. Concomitantly, a significant improvement of resilience, associated with increased leaf thickness and specific biochemical responses, was observed in defective NPR3 lines against E. necator and Plasmopara viticola. Transcriptomic analysis revealed that both MLO6-7 and NPR3 defective lines modulated their gene expression profiles, pointing to distinct though partially overlapping responses. Furthermore, targeted metabolite analysis highlighted an overaccumulation of stilbenes coupled with an improved oxidative scavenging potential in both editing targets, likely protecting the MLO6-7 mutants from detrimental pleiotropic effects. Finally, the Cre-loxP approach allowed the recovery of one MLO6-7 edited plant with the complete removal of transgene. Taken together, our achievements provide a comprehensive understanding of the molecular and biochemical adjustments occurring in double MLO-defective grape plants. In parallel, the potential of NPR3 mutants for multiple purposes has been demonstrated, raising new questions on its wide role in orchestrating biotic stress responses.
Keywords: Erysiphe necator; New Plant Breeding Techniques (NPBTs); Plasmopara viticola; Vitis vinifera; stilbenes.
© 2024 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interests. This article does not contain any studies with human or animal participants.
Figures
References
-
- Acevedo‐Garcia, J. , Kusch, S. & Panstruga, R. (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytologist, 204, 273–281. - PubMed
-
- Agliassa, C. , Mannino, G. , Molino, D. , Cavalletto, S. , Contartese, V. , Bertea, C.M. et al. (2021) A new protein hydrolysate‐based biostimulant applied by fertigation promotes relief from drought stress in Capsicum annuum L. Plant Physiology and Biochemistry, 166, 1076–1086. - PubMed
-
- Amarowicz, R. , Narolewska, O. , Karamac, M. , Kosinska, A. & Weidner, S. (2008) Grapevine leaves as a source of natural antioxidants. Polish Journal of Food and Nutrition Sciences, 58, 73–78.
-
- Anderson, R. , Bayer, P.E. & Edwards, D. (2020) Climate change and the need for agricultural adaptation. Current Opinion in Plant Biology, 56, 197–202. - PubMed
-
- Assink Junior, E.J. , Jesus, P.C.d. & Borges, E.M. (2023) Whey protein analysis using the Lowry assay and 96‐well‐plate digital images acquired using smartphones. Journal of Chemical Education, 100, 2329–2338.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

