Acetyltransferase NAT10 promotes an immunosuppressive microenvironment by modulating CD8+ T cell activity in prostate cancer
- PMID: 39648231
- PMCID: PMC11625704
- DOI: 10.1186/s43556-024-00228-5
Acetyltransferase NAT10 promotes an immunosuppressive microenvironment by modulating CD8+ T cell activity in prostate cancer
Abstract
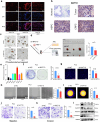
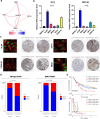
N-acetyltransferase 10 (NAT10), an enzyme responsible for ac4C acetylation, is implicated in cancer progression, though its specific biological function in prostate cancer remains insufficiently understood. This study clarifies NAT10's role in prostate cancer and its effects on the tumor immune microenvironment. NAT10 expression and clinical relevance were assessed through bioinformatics, RT-qPCR, and IHC analyses, comparing prostate cancer tissues with normal controls. The impact of NAT10 on tumor cell proliferation, migration, and invasion was investigated via in vitro assays-including CCK-8, EdU, wound healing, and 3D-Transwell-as well as in vivo mouse xenograft models and organoid studies. Further, NAT10's influence on immune cell infiltration was examined using flow cytometry, IHC, cell co-culture assays, and ELISA to elucidate downstream chemokine effects, specifically targeting CD8+ T cells. Findings indicated significant upregulation of NAT10 in prostate cancer cells, enhancing their proliferative and invasive capacities. Notably, NAT10 suppresses CD8+ T cell recruitment and cytotoxicity through the CCL25/CCR9 axis, fostering an immunosuppressive microenvironment that exacerbates tumor progression. An ac4C modification score was also devised based on NAT10's downstream targets, providing a novel predictive tool for evaluating immune infiltration and forecasting immunotherapy responses in patients with prostate cancer. This study underscores NAT10's pivotal role in modulating the prostate cancer immune microenvironment, offering insights into the immune desert phenomenon and identifying NAT10 as a promising therapeutic target for improving immunotherapy efficacy.
Keywords: Ac4C acetylation; Immune microenvironment; Immunotherapy; Prostate cancer.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study protocol was reviewed and approved by the Ethics Committee of [the ethics committee of Shanghai Tenth People's Hospital] (No:SHSY-IEC-5.0/19K110/P01). All participants provided written informed consent prior to inclusion in the study, and all procedures were conducted in accordance with the ethical principles outlined in the Declaration of Helsinki. For participants under the age of 18, consent was obtained from a parent or legal guardian. Data confidentiality and participant privacy were rigorously maintained throughout the study. Consent for publication: The authors have obtained written informed consent from all participants for the publication of their data and images included in this article. No identifiable personal information is included, and all data have been anonymized to ensure confidentiality. Competing interests: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. 10.3322/caac.21763. - PubMed
-
- Kang J, La Manna F, Bonollo F, Sampson N, Alberts IL, Mingels C, et al. Tumor microenvironment mechanisms and bone metastatic disease progression of prostate cancer. Cancer Lett. 2022;530:156–69. 10.1016/j.canlet.2022.01.015. - PubMed
-
- Chen XH, Guo KX, Li J, Xu SH, Zhu H, Yan GR. Regulations of m(6)A and other RNA modifications and their roles in cancer. Front Med. 2024;18(4):622–48. 10.1007/s11684-024-1064-8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
